

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
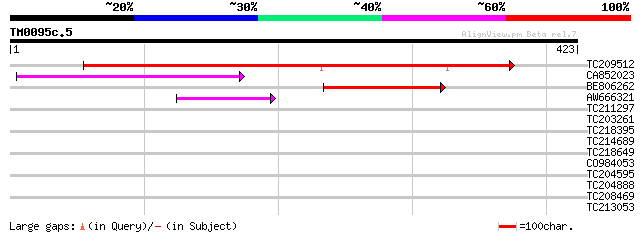
Query= TM0095c.5
(423 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209512 similar to UP|Q6SF22 (Q6SF22) Tetracycline resistance p... 494 e-140
CA852023 114 8e-26
BE806262 92 3e-19
AW666321 58 7e-09
TC211297 homologue to UP|Q9LFK1 (Q9LFK1) Sugar transporter-like ... 33 0.19
TC203261 similar to UP|Q84UU0 (Q84UU0) Photosystem I subunit XI,... 31 1.2
TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like pr... 30 1.6
TC214689 homologue to UP|Q40359 (Q40359) Alfin-1, partial (95%) 30 2.7
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 29 3.5
CO984053 28 6.0
TC204595 homologue to UP|CB21_CUCSA (P08221) Chlorophyll a-b bin... 28 6.0
TC204888 similar to UP|Q9ZTX0 (Q9ZTX0) Similarity to SCAMP37, pa... 28 6.0
TC208469 weakly similar to UP|Q9LIF0 (Q9LIF0) Similarity to phot... 28 7.8
TC213053 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter... 28 7.8
>TC209512 similar to UP|Q6SF22 (Q6SF22) Tetracycline resistance protein,
partial (5%)
Length = 1375
Score = 494 bits (1272), Expect = e-140
Identities = 262/355 (73%), Positives = 284/355 (79%), Gaps = 34/355 (9%)
Frame = +3
Query: 56 NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYV 115
NG+QQTIVGIFKMVVLPLLGQLSDE+GRKPLLLIT+ST IF F +L W QSEE+VYAYYV
Sbjct: 18 NGVQQTIVGIFKMVVLPLLGQLSDEYGRKPLLLITISTAIFPFVLLVWHQSEEYVYAYYV 197
Query: 116 LRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYI 175
LRT S IISQGSIFCISVAY ADVVNESKRAAVF WITGL SASHV+G+VLA LPE+YI
Sbjct: 198 LRTISNIISQGSIFCISVAYAADVVNESKRAAVFGWITGLLSASHVLGDVLAWSLPEKYI 377
Query: 176 FVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEI---- 231
F VSI LLT CPVYM+FFLVETV PAPK ++ SG K+VDV +RY SMR AAEI
Sbjct: 378 FAVSIVLLTSCPVYMKFFLVETVIPAPKNDRESGCWAKIVDVPRQRYISMRRAAEIVIFS 557
Query: 232 ----------------LDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPI 275
+ + + YYLKAVFGFNKNQFSELLMMVGIGSIFSQM+LLPI
Sbjct: 558 PTLRGMALVSFFYELGMSGISSVLLYYLKAVFGFNKNQFSELLMMVGIGSIFSQMLLLPI 737
Query: 276 LNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS---------- 325
LNP VGEKVIL +ALLASIAYAWLYGLAWAPWVPY SASFGIIYVLVKP+
Sbjct: 738 LNPLVGEKVILCSALLASIAYAWLYGLAWAPWVPYLSASFGIIYVLVKPATYAIISNASS 917
Query: 326 ----GKAQTFIAGAQSISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
GKAQTFIAG QSISDLLSPIAMSPLTSWFLSSNAPF+CKGFSII AS+ M+
Sbjct: 918 STNQGKAQTFIAGTQSISDLLSPIAMSPLTSWFLSSNAPFECKGFSIISASICMI 1082
>CA852023
Length = 827
Score = 114 bits (285), Expect = 8e-26
Identities = 59/170 (34%), Positives = 95/170 (55%)
Frame = +3
Query: 6 GLIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPGGSTCPKVIYINGLQQTIVGI 65
G+ L L HL + + + + + DVT +ALCPG C IY++G QQ + G+
Sbjct: 105 GMEGLGGLGHLFVTMFVTGFGGVIVLPAITDVTMAALCPGQDQCSLAIYLSGFQQAMAGV 284
Query: 66 FKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQ 125
+V+ PL+G LSD +GRK LL + ++ ++ +LA+ + +F YAYYV +T + + +
Sbjct: 285 GSVVMTPLIGNLSDRYGRKALLTLPLTVSVIPQVILAYSRDTQFFYAYYVGKTLAAMAGE 464
Query: 126 GSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYI 175
GS C+++AYVAD V + KR + G + + + ARFL YI
Sbjct: 465 GSFHCLALAYVADKVPDGKRHQHSEYFAGGWISIVRWWTLAARFLSTAYI 614
>BE806262
Length = 417
Score = 92.4 bits (228), Expect = 3e-19
Identities = 43/91 (47%), Positives = 60/91 (65%)
Frame = +3
Query: 235 LIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASI 294
L+ + YYLKA F FNKNQF++LLM+ GIG+ +Q+ +PIL P +GE+ +LS LL S
Sbjct: 126 LMAVLLYYLKARFQFNKNQFADLLMITGIGATLAQLFFMPILVPVIGEEKLLSTGLLISC 305
Query: 295 AYAWLYGLAWAPWVPYFSASFGIIYVLVKPS 325
++Y +AW WVPY A + V V+PS
Sbjct: 306 INVFVYSIAWTAWVPYALAGCSVFAVFVRPS 398
>AW666321
Length = 422
Score = 58.2 bits (139), Expect = 7e-09
Identities = 31/74 (41%), Positives = 44/74 (58%)
Frame = +1
Query: 125 QGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLT 184
+GS C+++AYVAD V + KR + F + G+ SAS V G + ARFL F V+ L
Sbjct: 13 EGSFHCLALAYVADKVPDGKRTSAFGILAGVGSASFVGGTLAARFLSTALTFQVASVLSM 192
Query: 185 FCPVYMQFFLVETV 198
VYM+ FL ++V
Sbjct: 193 VALVYMRIFLKDSV 234
>TC211297 homologue to UP|Q9LFK1 (Q9LFK1) Sugar transporter-like protein,
partial (8%)
Length = 817
Score = 33.5 bits (75), Expect = 0.19
Identities = 14/35 (40%), Positives = 24/35 (68%)
Frame = +1
Query: 234 NLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFS 268
++ + VQY+LKA F F+KNQ ++L+++ I S
Sbjct: 58 HIFLRVQYFLKAQFHFDKNQLADLMVIFEIAGTVS 162
>TC203261 similar to UP|Q84UU0 (Q84UU0) Photosystem I subunit XI, partial
(88%)
Length = 997
Score = 30.8 bits (68), Expect = 1.2
Identities = 35/153 (22%), Positives = 62/153 (39%)
Frame = +1
Query: 205 NQVSGFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIG 264
N++ G T +LH K + + L+ L+ + +++ VF Q + ++M
Sbjct: 43 NEIGGASTATTLILH*AMKLLSHLGSSLETLVY*L-FFVVVVFHL*N*QQHKAIIMAAAS 219
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
+ SQ+ K + L+A + L P FS + I KP
Sbjct: 220 PMASQL------------KSTFTRTLVAPKGLSASSPLHLVPSRRQFSFTVKAIQS-EKP 360
Query: 325 SGKAQTFIAGAQSISDLLSPIAMSPLTSWFLSS 357
+ + I G I L +P+ SPL +W+LS+
Sbjct: 361 TYQVIQPINGDPFIGSLETPVTSSPLIAWYLSN 459
>TC218395 similar to UP|Q9FIF2 (Q9FIF2) Sugar transporter-like protein,
partial (40%)
Length = 1013
Score = 30.4 bits (67), Expect = 1.6
Identities = 37/153 (24%), Positives = 69/153 (44%), Gaps = 17/153 (11%)
Frame = +1
Query: 62 IVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYY------- 114
++G+FK+++ + D+ GR+PLL+ +S S +L+ AYY
Sbjct: 253 VIGLFKLLMTWIAVLKVDDLGRRPLLIGGVSGIALSLVLLS---------AYYKFLGGFP 405
Query: 115 -------VLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVG---N 164
+L Y IS G I + V+ V + K ++ + F+++ VV +
Sbjct: 406 LVAVGALLLYVGCYQISFGPISWLMVSEVFPLRTRGKGISLA--VLTNFASNAVVTFAFS 579
Query: 165 VLARFLPEEYIFVVSIALLTFCPVYMQFFLVET 197
L FL E +F++ A+ T +++ F + ET
Sbjct: 580 PLKEFLGAENLFLLFGAIATLSLLFIIFSVPET 678
>TC214689 homologue to UP|Q40359 (Q40359) Alfin-1, partial (95%)
Length = 1352
Score = 29.6 bits (65), Expect = 2.7
Identities = 12/29 (41%), Positives = 18/29 (61%)
Frame = +1
Query: 233 DNLIIAVQYYLKAVFGFNKNQFSELLMMV 261
D+ ++AV +Y A FGF KN+ L M+
Sbjct: 508 DSWLLAVAFYFGARFGFGKNERKRLFQMI 594
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 29.3 bits (64), Expect = 3.5
Identities = 38/197 (19%), Positives = 76/197 (38%)
Frame = +2
Query: 75 GQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVA 134
GQ+++ GRK L+I I + +++ + F+Y +L F G I
Sbjct: 347 GQIAEYIGRKGSLMIASIPNIIGWLAISFAKDSSFLYMGRLLEGFGV----GIISYTVPV 514
Query: 135 YVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPVYMQFFL 194
Y+A++ + R + S + ++ +L F+ + ++ I T + FF+
Sbjct: 515 YIAEISPPNLRGGLVSVNQLSVTIGIMLAYLLGIFVEWRILAIIGILPCTIL-IPALFFI 691
Query: 195 VETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQYYLKAVFGFNKNQF 254
E+ K F T + VL + + + + + F K +
Sbjct: 692 PESPRWLAKMGMTEEFETS-LQVLRGFDTDISVEVNEIKRAVASTNTRITVRFADLKQRR 868
Query: 255 SELLMMVGIGSIFSQMV 271
L +M+GIG + Q +
Sbjct: 869 YWLPLMIGIGLLILQQL 919
>CO984053
Length = 880
Score = 28.5 bits (62), Expect = 6.0
Identities = 11/46 (23%), Positives = 26/46 (55%)
Frame = -3
Query: 16 LLFPLSIHWIAEEMTVSVLVDVTTSALCPGGSTCPKVIYINGLQQT 61
+L + +H++++ M +L D+ SA G + P ++Y+ + +T
Sbjct: 605 ILERIVVHYVSQRMAWKLLKDIPRSATRKAGRSMPTLVYVYCVSRT 468
>TC204595 homologue to UP|CB21_CUCSA (P08221) Chlorophyll a-b binding protein
of LHCII type I, chloroplast precursor (CAB) (LHCP)
(Fragment), partial (30%)
Length = 1279
Score = 28.5 bits (62), Expect = 6.0
Identities = 24/84 (28%), Positives = 38/84 (44%), Gaps = 4/84 (4%)
Frame = +1
Query: 241 YYLKA--VFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSA--ALLASIAY 296
YYL A ++ +K + L M + + + FS V +P+ +K SA LL A
Sbjct: 295 YYLHAKHMYNLDKMKHQGLKMSLAVFTAFSIGVAVPVYAVIFQQKKTASA*AHLLLLFAI 474
Query: 297 AWLYGLAWAPWVPYFSASFGIIYV 320
W++G + V F + IYV
Sbjct: 475 RWIWGF*TSLLVLVFITALNFIYV 546
>TC204888 similar to UP|Q9ZTX0 (Q9ZTX0) Similarity to SCAMP37, partial (46%)
Length = 557
Score = 28.5 bits (62), Expect = 6.0
Identities = 23/70 (32%), Positives = 32/70 (44%), Gaps = 10/70 (14%)
Frame = +1
Query: 105 QSEEFVYAYYVLRTFS----------YIISQGSIFCISVAYVADVVNESKRAAVFSWITG 154
+SE V YY+L + + YI+ G FCI A +V + K +TG
Sbjct: 55 RSENMVSCYYLLHSRNSWSMSCGIAHYIVLLG--FCILAAVAPPIVFKGKS------LTG 210
Query: 155 LFSASHVVGN 164
+ SA VVGN
Sbjct: 211 ILSAIDVVGN 240
>TC208469 weakly similar to UP|Q9LIF0 (Q9LIF0) Similarity to
photoreceptor-interacting transcription factor, partial
(20%)
Length = 847
Score = 28.1 bits (61), Expect = 7.8
Identities = 15/41 (36%), Positives = 25/41 (60%)
Frame = +1
Query: 368 IICASVSMVAETENEKLKENLQDEKNKNIELEKQMIKLKAQ 408
++ S + T+NEKLK +LQ + + +ELEK K++ Q
Sbjct: 481 VVLYSSNFDISTDNEKLKAHLQGMQWRVMELEKFCRKMQIQ 603
>TC213053 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(27%)
Length = 440
Score = 28.1 bits (61), Expect = 7.8
Identities = 16/44 (36%), Positives = 22/44 (49%)
Frame = +1
Query: 58 LQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVL 101
L VG K V + + L D GR+PLLL ++ +FS L
Sbjct: 61 LATVAVGFAKTVFILVATFLLDRVGRRPLLLTSVGGMVFSLLTL 192
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,142,115
Number of Sequences: 63676
Number of extensions: 280617
Number of successful extensions: 2071
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 2056
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2068
length of query: 423
length of database: 12,639,632
effective HSP length: 100
effective length of query: 323
effective length of database: 6,272,032
effective search space: 2025866336
effective search space used: 2025866336
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0095c.5


