

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093b.6
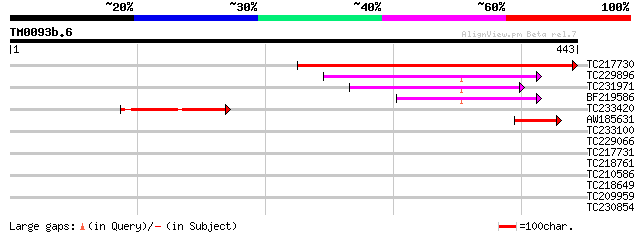
(443 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217730 similar to GB|AAQ56808.1|34098851|BT010365 At5g07720 {A... 447 e-126
TC229896 similar to UP|Q9ST56 (Q9ST56) Alpha galactosyltransfera... 152 4e-37
TC231971 similar to UP|Q9ST56 (Q9ST56) Alpha galactosyltransfera... 113 1e-25
BF219586 similar to PIR|T52082|T52 alpha galactosyltransferase (... 91 1e-18
TC233420 similar to UP|Q7X9N4 (Q7X9N4) Galactomannan galactosylt... 86 3e-17
AW185631 43 3e-04
TC233100 homologue to UP|Q40202 (Q40202) RAB1B (Fragment), parti... 41 0.001
TC229066 weakly similar to GB|AAO24542.1|27808524|BT003110 At1g5... 32 0.44
TC217731 32 0.44
TC218761 similar to UP|Q9XIS4 (Q9XIS4) Starch branching enzyme ... 31 1.3
TC210586 homologue to UP|P2A_TOBAC (Q9XGH7) Serine/threonine pro... 29 4.9
TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein... 28 6.3
TC209959 weakly similar to UP|Q9ZQ45 (Q9ZQ45) Expressed protein,... 28 6.3
TC230854 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g3... 28 8.3
>TC217730 similar to GB|AAQ56808.1|34098851|BT010365 At5g07720 {Arabidopsis
thaliana;} , partial (46%)
Length = 950
Score = 447 bits (1151), Expect = e-126
Identities = 208/218 (95%), Positives = 216/218 (98%)
Frame = +3
Query: 226 PLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREE 285
P+SKYD+YNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREE
Sbjct: 6 PMSKYDEYNLVLHGYPDLLFEQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREE 185
Query: 286 AGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEM 345
AGK+LTANLKGRPAFEADDQSALIYLLLSKK+KWMDKTFLENSFYLHGYWAGLVDRYEEM
Sbjct: 186 AGKILTANLKGRPAFEADDQSALIYLLLSKKEKWMDKTFLENSFYLHGYWAGLVDRYEEM 365
Query: 346 IEKYHPGLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFNFADNQVLKLYGFRHR 405
IEKYHPGLGDERWPFVTHFVGCKPCGSYGDYPVE+CLSSMERAFNFADNQVLKLYGFRHR
Sbjct: 366 IEKYHPGLGDERWPFVTHFVGCKPCGSYGDYPVERCLSSMERAFNFADNQVLKLYGFRHR 545
Query: 406 GLLSPKIKRIRNETVTPLEFVDQFDIRRHSSESRGSKS 443
GLLSPKIKRIRNETV+PLEFVDQFDIRRHS+E+ SKS
Sbjct: 546 GLLSPKIKRIRNETVSPLEFVDQFDIRRHSTENTESKS 659
>TC229896 similar to UP|Q9ST56 (Q9ST56) Alpha galactosyltransferase
(Fragment) , partial (43%)
Length = 1091
Score = 152 bits (383), Expect = 4e-37
Identities = 75/205 (36%), Positives = 111/205 (53%), Gaps = 35/205 (17%)
Frame = +3
Query: 246 EQKSWIAVNTGSFLFRNCQWSLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQ 305
+++SW +N G FL RNCQWSLD ++AWA MGP+ P E+ G+ L + K + E+DDQ
Sbjct: 3 QKRSWTGLNAGVFLIRNCQWSLDFMEAWASMGPQTPNYEKWGQTLRSTFKDKFFPESDDQ 182
Query: 306 SALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHP-------------- 351
+ L YL+ +KDKW D+ +LE+ +Y GYW ++ ++ + EKY+
Sbjct: 183 TGLAYLIAIEKDKWADRIYLESEYYFEGYWEEILGTFQNITEKYNEMEKGVSRLRRRHAE 362
Query: 352 -------------------GLGDERWPFVTHFVGCKPC-GSYGD-YPVEKCLSSMERAFN 390
G G R PF+THF GC+PC G Y Y + C + M++A N
Sbjct: 363 KVSETYGEMREEYLKDAGNGKGSWRRPFITHFTGCQPCSGKYNAMYSADDCWNGMQKALN 542
Query: 391 FADNQVLKLYGFRHRGLLSPKIKRI 415
FADNQV++ +G+ LL I +
Sbjct: 543 FADNQVMRKFGYMRPDLLDNAISPV 617
>TC231971 similar to UP|Q9ST56 (Q9ST56) Alpha galactosyltransferase
(Fragment) , partial (35%)
Length = 728
Score = 113 bits (283), Expect = 1e-25
Identities = 59/172 (34%), Positives = 87/172 (50%), Gaps = 35/172 (20%)
Frame = +1
Query: 266 SLDLLDAWAPMGPKGPVREEAGKVLTANLKGRPAFEADDQSALIYLLLSKKDKWMDKTFL 325
SLD ++AWA MGP+ P E+ G+ L + K + E+DDQ+ L YL+ +KDKW ++ +L
Sbjct: 1 SLDFMEAWASMGPQSPNYEKWGQTLRSTFKDKFFPESDDQTGLAYLIAMEKDKWAERIYL 180
Query: 326 ENSFYLHGYWAGLVDRYEEMIEKYHP---------------------------------G 352
E+ +Y GYW + ++ + EKY
Sbjct: 181 ESEYYFEGYWEEIQGTFKNITEKYKEMEKGVQRLRRRHAEKVSETYGEMREEYLKDAGNA 360
Query: 353 LGDERWPFVTHFVGCKPC-GSYGD-YPVEKCLSSMERAFNFADNQVLKLYGF 402
G R PF+THF GC+PC G Y Y C ++M A NFADNQV++ +G+
Sbjct: 361 KGSWRRPFITHFTGCQPCSGKYNAMYSAHDCWNAMHNALNFADNQVMRKFGY 516
>BF219586 similar to PIR|T52082|T52 alpha galactosyltransferase (EC 2.4.1.-)
[imported] - Trigonella foenum-graecum (fragment),
partial (30%)
Length = 679
Score = 90.5 bits (223), Expect = 1e-18
Identities = 48/148 (32%), Positives = 69/148 (46%), Gaps = 35/148 (23%)
Frame = -2
Query: 303 DDQSALIYLLLSKKDKWMDKTFLENSFYLHGYWAGLVDRYEEMIEKYHP----------- 351
DDQ+ L Y +KDKW D+ +LE+ +Y GYW ++ + EKY+
Sbjct: 666 DDQTGLAYXXXIEKDKWADRIYLESEYYFEGYWEEXXGTFQNITEKYNEMEKGVSRLRRR 487
Query: 352 ----------------------GLGDERWPFVTHFVGCKPC-GSYGD-YPVEKCLSSMER 387
G G R PF+THF GC+PC G Y Y + C + M++
Sbjct: 486 HAEKVSETYGEMREEYLKDAGNGKGSWRRPFITHFTGCQPCSGKYNAMYSADDCWNGMQK 307
Query: 388 AFNFADNQVLKLYGFRHRGLLSPKIKRI 415
A NFADNQV++ +G+ LL I +
Sbjct: 306 ALNFADNQVMRKFGYMRPDLLDNAISPV 223
>TC233420 similar to UP|Q7X9N4 (Q7X9N4) Galactomannan galactosyltransferase,
partial (18%)
Length = 466
Score = 85.9 bits (211), Expect = 3e-17
Identities = 41/87 (47%), Positives = 54/87 (61%), Gaps = 1/87 (1%)
Frame = +3
Query: 87 DPDDAAAAETFFS-PNATFTLGPKITGWDLQRKAWLDQNPEYPNFVRGKARILLLTGSPP 145
DP D TF+ P +T+ K+ WD +R+ WL +P + R R+ ++TGS P
Sbjct: 219 DPPD----RTFYDDPQMGYTMDKKVRNWDEKREEWLKLHPSFAAGAR--ERVFMVTGSQP 380
Query: 146 KPCDNPIGDHYLLKSIKNKIDYCRLHG 172
KPC NPIGDH LL+ KNK+DYCRLHG
Sbjct: 381 KPCRNPIGDHLLLRFFKNKVDYCRLHG 461
>AW185631
Length = 339
Score = 42.7 bits (99), Expect = 3e-04
Identities = 19/37 (51%), Positives = 25/37 (67%)
Frame = +2
Query: 395 QVLKLYGFRHRGLLSPKIKRIRNETVTPLEFVDQFDI 431
Q+L +YGF H+ L S +KRIRNET PLE D+ +
Sbjct: 2 QILHIYGFTHKSLGSRGVKRIRNETSNPLEVKDELGL 112
>TC233100 homologue to UP|Q40202 (Q40202) RAB1B (Fragment), partial (22%)
Length = 779
Score = 41.2 bits (95), Expect = 0.001
Identities = 19/35 (54%), Positives = 26/35 (74%)
Frame = -2
Query: 20 GGRRGRQIQKTFNNVKITILCGFVTILVLRGTIGV 54
G RR RQ+Q+ + +T LC F+T++VLRGTIGV
Sbjct: 256 GPRRVRQMQRACRHSTVTFLCLFLTLVVLRGTIGV 152
>TC229066 weakly similar to GB|AAO24542.1|27808524|BT003110 At1g56320
{Arabidopsis thaliana;} , partial (42%)
Length = 858
Score = 32.3 bits (72), Expect = 0.44
Identities = 20/70 (28%), Positives = 38/70 (53%), Gaps = 2/70 (2%)
Frame = +1
Query: 200 MLSHPEVEWIWWMDSDAF--FTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGS 257
+LSH EV + + + +F F+D+++ P+ KY+ V+ D + ++W S
Sbjct: 343 VLSHIEVSSLTAIATSSFNCFSDVLYGCPVVKYEVAENVVQAIEDQVCL*QTWSLAECIS 522
Query: 258 FLFRNCQWSL 267
FL+R+ Q S+
Sbjct: 523 FLYRHIQVSI 552
>TC217731
Length = 410
Score = 32.3 bits (72), Expect = 0.44
Identities = 14/19 (73%), Positives = 16/19 (83%)
Frame = +3
Query: 424 EFVDQFDIRRHSSESRGSK 442
EFVDQFDIRRHS+E+ K
Sbjct: 30 EFVDQFDIRRHSTENTXIK 86
>TC218761 similar to UP|Q9XIS4 (Q9XIS4) Starch branching enzyme , partial
(12%)
Length = 780
Score = 30.8 bits (68), Expect = 1.3
Identities = 20/58 (34%), Positives = 30/58 (51%)
Frame = +2
Query: 52 IGVNLSSSDADAVNQNVIEETNRILAEIRSDADPSDPDDAAAAETFFSPNATFTLGPK 109
I L S + + +N + +EET + A + S+ P+DAA E+FF N LG K
Sbjct: 431 ISGELVSVETEGINLDKLEETI-VAASVESEVVRDKPEDAAN*ESFFLVNIKCFLGFK 601
>TC210586 homologue to UP|P2A_TOBAC (Q9XGH7) Serine/threonine protein
phosphatase PP2A catalytic subunit , partial (30%)
Length = 840
Score = 28.9 bits (63), Expect = 4.9
Identities = 16/60 (26%), Positives = 28/60 (46%)
Frame = -1
Query: 200 MLSHPEVEWIWWMDSDAFFTDMVFELPLSKYDDYNLVLHGYPDLLFEQKSWIAVNTGSFL 259
++SH +V IW + S+A + ELP++ + N H LF Q + + +L
Sbjct: 441 IISHKQVICIWTLPSNAKKLCKIMELPMNITTNCNRTFHRLYIALFHQNLFCLITQDPYL 262
>TC218649 similar to UP|Q39416 (Q39416) Integral membrane protein, partial
(93%)
Length = 1695
Score = 28.5 bits (62), Expect = 6.3
Identities = 15/39 (38%), Positives = 16/39 (40%)
Frame = -3
Query: 352 GLGDERWPFVTHFVGCKPCGSYGDYPVEKCLSSMERAFN 390
G G R P H C C G PV KC + AFN
Sbjct: 1393 GEGSSRAPIQKHI--CCQCNQPGKQPVGKCCHTSREAFN 1283
>TC209959 weakly similar to UP|Q9ZQ45 (Q9ZQ45) Expressed protein, partial
(28%)
Length = 632
Score = 28.5 bits (62), Expect = 6.3
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Frame = +2
Query: 56 LSSSDADAVNQNVIEETNRILAEIRSDADPSDPDD--AAAAETFFSPNATFTLGP 108
+SSS + V V E + LA I+S DPS P D +A E F NA +GP
Sbjct: 275 ISSSKSQEVVDQVGREIEQALATIQSPHDPSAPVDQKSALTELKFKLNA---IGP 430
>TC230854 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g31350
{Arabidopsis thaliana;} , partial (12%)
Length = 667
Score = 28.1 bits (61), Expect = 8.3
Identities = 15/51 (29%), Positives = 23/51 (44%), Gaps = 5/51 (9%)
Frame = -1
Query: 149 DNPIGDHYLLKSIK-----NKIDYCRLHGIEIVYNLAHLDVELAGYWAKLP 194
D+ GDH +L + N ++YCR+ LD + +W KLP
Sbjct: 265 DSEAGDHMVLDHQRMMRNINTLEYCRIR--------VELDQSILSFWEKLP 137
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,902,452
Number of Sequences: 63676
Number of extensions: 340645
Number of successful extensions: 1378
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 1366
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1374
length of query: 443
length of database: 12,639,632
effective HSP length: 100
effective length of query: 343
effective length of database: 6,272,032
effective search space: 2151306976
effective search space used: 2151306976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0093b.6


