

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093a.1
(268 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
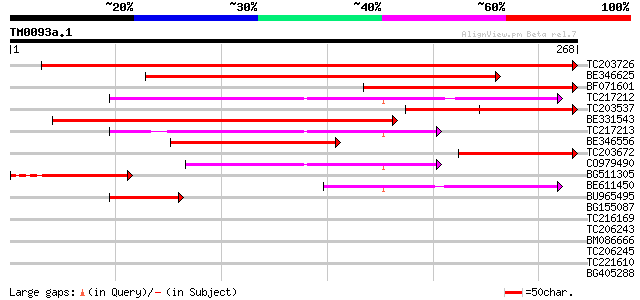
Score E
Sequences producing significant alignments: (bits) Value
TC203726 homologue to UP|Q8S4X2 (Q8S4X2) Ribulose-5-phosphate-3-... 474 e-134
BE346625 similar to GP|19071636|gb ribulose-5-phosphate-3-epimer... 222 1e-58
BF071601 similar to SP|Q43843|RPE_ Ribulose-phosphate 3-epimeras... 167 5e-42
TC217212 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-... 157 4e-39
TC203537 homologue to UP|RPE_SOLTU (Q43843) Ribulose-phosphate 3... 154 3e-38
BE331543 122 2e-28
TC217213 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-... 115 3e-26
BE346556 weakly similar to GP|4930130|pdb| Chain A D-Ribulose-5... 114 5e-26
TC203672 homologue to UP|Q8S4X2 (Q8S4X2) Ribulose-5-phosphate-3-... 107 7e-24
CO979490 91 7e-19
BG511305 similar to GP|20339362|gb| ribulose-5-phosphate-3-epime... 78 4e-15
BE611450 similar to GP|6007803|gb|A D-ribulose-5-phosphate 3-epi... 58 5e-09
BU965495 41 5e-04
BG155087 36 0.021
TC216169 weakly similar to UP|Q6JJ29 (Q6JJ29) Prephenate dehydra... 29 1.9
TC206243 similar to PIR|B86314|B86314 F2H15.12 protein - Arabido... 29 2.5
BM086666 similar to SP|O82660|H136_ Photosystem II stability/ass... 29 2.5
TC206245 similar to UP|O49213 (O49213) GDP-4-keto-6-deoxy-D-mann... 29 2.5
TC221610 28 3.3
BG405288 homologue to GP|20339362|gb| ribulose-5-phosphate-3-epi... 28 4.3
>TC203726 homologue to UP|Q8S4X2 (Q8S4X2) Ribulose-5-phosphate-3-epimerase,
complete
Length = 1748
Score = 474 bits (1219), Expect = e-134
Identities = 241/253 (95%), Positives = 247/253 (97%)
Frame = +3
Query: 16 NHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWI 75
+H SLTFSRRKIST VKA+SRVDKFSKSDIIVSPSILSANF+KLGEQVKAVELAGCDWI
Sbjct: 573 SHTPSLTFSRRKISTTVKATSRVDKFSKSDIIVSPSILSANFSKLGEQVKAVELAGCDWI 752
Query: 76 HVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCE 135
HVDVMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVS+HCE
Sbjct: 753 HVDVMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCE 932
Query: 136 QSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIE 195
QSSTIHLHRTVNQVKSLGAKAGVVLNP TPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIE
Sbjct: 933 QSSTIHLHRTVNQVKSLGAKAGVVLNPATPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIE 1112
Query: 196 SQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAI 255
SQVKKISDLRRLCAEKGVNPWIEVDGGV PANAYKVIEAGANALVAGSAVFGAKDYAEAI
Sbjct: 1113SQVKKISDLRRLCAEKGVNPWIEVDGGVGPANAYKVIEAGANALVAGSAVFGAKDYAEAI 1292
Query: 256 KGIKTSKRPEPVA 268
+GIKTSKRPE VA
Sbjct: 1293RGIKTSKRPEVVA 1331
>BE346625 similar to GP|19071636|gb ribulose-5-phosphate-3-epimerase {Oryza
sativa (japonica cultivar-group)}, partial (56%)
Length = 505
Score = 222 bits (566), Expect = 1e-58
Identities = 117/168 (69%), Positives = 131/168 (77%)
Frame = +2
Query: 65 KAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIK 124
KA+ELAGCD IHVD+ D R V NITIGPL+VDAL PVT LPL VHLMIV+PEQRVP FI
Sbjct: 2 KALELAGCDRIHVDITDRRVVANITIGPLVVDALHPVTHLPLHVHLMIVDPEQRVPYFIN 181
Query: 125 AGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSV 184
AG D+VS+HCEQSST HLH TVN VK+LGAK GVVL P T SA++YVLDV DLVLIM
Sbjct: 182 AGPDLVSVHCEQSSTNHLHGTVN*VKNLGAKPGVVLYPATHYSALDYVLDVEDLVLIMYR 361
Query: 185 NPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVI 232
NPG GGQ IESQV K+SD+RR+CA GVN IEV G A+ + VI
Sbjct: 362 NPGCGGQRCIESQVHKMSDIRRVCARYGVNTCIEVHCGWRAAHVHTVI 505
>BF071601 similar to SP|Q43843|RPE_ Ribulose-phosphate 3-epimerase
chloroplast precursor (EC 5.1.3.1) (Pentose-5-phosphate
3-epimerase), partial (36%)
Length = 404
Score = 167 bits (423), Expect = 5e-42
Identities = 85/101 (84%), Positives = 89/101 (87%)
Frame = +3
Query: 168 AIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPAN 227
AIEY+LDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGV P N
Sbjct: 3 AIEYILDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVGPXN 182
Query: 228 AYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
A V AGA AL AGSAV GA DYA++I+GIKT KRPE VA
Sbjct: 183 AIPVTGAGATALAAGSAVIGANDYAKSIRGIKTIKRPEVVA 305
>TC217212 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-epimerase ,
partial (94%)
Length = 843
Score = 157 bits (398), Expect = 4e-39
Identities = 86/217 (39%), Positives = 130/217 (59%), Gaps = 3/217 (1%)
Frame = +3
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + + + G DW+H+D+MDG FVPN+TIG ++++LR T LD
Sbjct: 57 IAPSMLSSDFANLASEAQRMLHFGADWLHMDIMDGHFVPNLTIGAPVIESLRKHTKGYLD 236
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLS 167
HLM+ P V KAGA + H E S + + ++KS G GV L PGTP+
Sbjct: 237 CHLMVTNPLDYVEPLAKAGASGFTFHVETSKD-NWEELIQRIKSHGMTPGVALKPGTPIE 413
Query: 168 AIEYVLDV---VDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVT 224
+ +++ V++VL+M+V PGFGGQ F+ + K+ LR+ K + IEVDGG+
Sbjct: 414 EVYPLVEAGNPVEMVLVMTVEPGFGGQKFMPEMMDKVRILRK----KYPSLDIEVDGGLG 581
Query: 225 PANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTS 261
P+ AGAN +VAGS+VFGA + + I ++ S
Sbjct: 582 PSTIDVAASAGANCIVAGSSVFGAPEPVQVISLLRNS 692
>TC203537 homologue to UP|RPE_SOLTU (Q43843) Ribulose-phosphate 3-epimerase,
chloroplast precursor (Pentose-5-phosphate 3-epimerase)
(PPE) (RPE) (R5P3E) (Fragment) , partial (29%)
Length = 715
Score = 154 bits (390), Expect = 3e-38
Identities = 77/81 (95%), Positives = 79/81 (97%)
Frame = -2
Query: 188 FGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFG 247
FGGQSFIESQVKKISDLRR+CAEKGVNPWIEVDGGV PANAYKVIEAGANALVAGSAVFG
Sbjct: 714 FGGQSFIESQVKKISDLRRVCAEKGVNPWIEVDGGVGPANAYKVIEAGANALVAGSAVFG 535
Query: 248 AKDYAEAIKGIKTSKRPEPVA 268
AKDYAEAI+GIKTSKRPE VA
Sbjct: 534 AKDYAEAIRGIKTSKRPEAVA 472
Score = 84.7 bits (208), Expect = 4e-17
Identities = 43/46 (93%), Positives = 44/46 (95%)
Frame = +2
Query: 223 VTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
V PANAYKVIEAGANALVAGSAVFGAKDYAEAI+GIKTSKRPE VA
Sbjct: 2 VGPANAYKVIEAGANALVAGSAVFGAKDYAEAIRGIKTSKRPEAVA 139
>BE331543
Length = 562
Score = 122 bits (305), Expect = 2e-28
Identities = 78/163 (47%), Positives = 104/163 (62%)
Frame = +2
Query: 21 LTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVM 80
LT SRRK+++ +K +SRVD SK DI +S +ILSA F K +Q A + IH+D+M
Sbjct: 74 LTLSRRKMASTLKDTSRVDPCSKCDITISMTILSAYFEK*EQQDNAKD*LC*Y*IHLDMM 253
Query: 81 DGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTI 140
G V ITI PL VDA PVTDL LD+H EPE +PD I A + I S+H EQS +I
Sbjct: 254 HGTIVSLITIIPLDVDAELPVTDLALDLHQKSGEPEHLIPDCI*ARSYIHSVH*EQSYSI 433
Query: 141 HLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMS 183
H+HRT N V S GA AG+ LSA++Y+L++ + V I++
Sbjct: 434 HVHRTDNHVISTGAIAGLDYIVRLLLSALQYILNMFNSVSIIT 562
>TC217213 similar to UP|Q9SE42 (Q9SE42) D-ribulose-5-phosphate 3-epimerase ,
partial (74%)
Length = 646
Score = 115 bits (287), Expect = 3e-26
Identities = 61/160 (38%), Positives = 95/160 (59%), Gaps = 3/160 (1%)
Frame = +1
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PS+LS++FA L + + + +H+D+MDG FVPN+TIG ++++LR T LD
Sbjct: 28 IAPSMLSSDFANLASEAQRM-------LHMDIMDGHFVPNLTIGAPVIESLRKHTKGYLD 186
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLS 167
HLM+ P V KAGA + H E S + + ++KS G GV L PGTP+
Sbjct: 187 CHLMVTNPLDYVEPLAKAGASGFTFHVETSKD-NWEELIQRIKSHGMTPGVALKPGTPIE 363
Query: 168 AIEYVLDV---VDLVLIMSVNPGFGGQSFIESQVKKISDL 204
+ +++ V++VL+M+V PGFGGQ F+ + K+S L
Sbjct: 364 EVYPLVEAGNPVEMVLVMTVEPGFGGQKFMPEMMDKVSSL 483
>BE346556 weakly similar to GP|4930130|pdb| Chain A D-Ribulose-5-Phosphate
3-Epimerase From Solanum Tuberosum Chloroplasts, partial
(34%)
Length = 241
Score = 114 bits (285), Expect = 5e-26
Identities = 53/80 (66%), Positives = 68/80 (84%)
Frame = +2
Query: 77 VDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQ 136
VDVMDGRFVPNITI PL++D L P+T+LPL +HL+I+ P+Q +P FIKA ++IV+IH EQ
Sbjct: 2 VDVMDGRFVPNITIEPLVIDTLHPLTNLPLHIHLIIINPKQIIPYFIKAKSNIVNIHYEQ 181
Query: 137 SSTIHLHRTVNQVKSLGAKA 156
SSTIHLH T+NQ+K+L KA
Sbjct: 182 SSTIHLHHTINQLKTLITKA 241
>TC203672 homologue to UP|Q8S4X2 (Q8S4X2) Ribulose-5-phosphate-3-epimerase,
partial (20%)
Length = 860
Score = 107 bits (266), Expect = 7e-24
Identities = 53/56 (94%), Positives = 54/56 (95%)
Frame = +1
Query: 213 VNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
VNPWIEVDGGV PANAYKVIEAGANALVAGSAVFGAKDYAEAI+GIKTSKRPE VA
Sbjct: 1 VNPWIEVDGGVGPANAYKVIEAGANALVAGSAVFGAKDYAEAIRGIKTSKRPEVVA 168
>CO979490
Length = 740
Score = 90.5 bits (223), Expect = 7e-19
Identities = 49/124 (39%), Positives = 72/124 (57%), Gaps = 3/124 (2%)
Frame = -2
Query: 84 FVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLH 143
FVPN+TIG ++++LR T LD HLM+ P V KAGA + H E S +
Sbjct: 571 FVPNLTIGAPVIESLRKHTKAYLDCHLMVTNPLDYVEPLAKAGASGFTFHVETSKD-NWK 395
Query: 144 RTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDV---VDLVLIMSVNPGFGGQSFIESQVKK 200
+ ++KS G GV L PGTP+ + +++ V++VL+M+V PGFGGQ F+ + K
Sbjct: 394 ELIQRIKSHGMIPGVALKPGTPVGEVYPLVEAENPVEMVLVMTVEPGFGGQKFMAETMDK 215
Query: 201 ISDL 204
+S L
Sbjct: 214 VSSL 203
>BG511305 similar to GP|20339362|gb| ribulose-5-phosphate-3-epimerase {Pisum
sativum}, partial (23%)
Length = 421
Score = 78.2 bits (191), Expect = 4e-15
Identities = 46/58 (79%), Positives = 51/58 (87%)
Frame = +2
Query: 1 INGPSSLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFA 58
ING LH KT++ +HPRSLTFSR+KIST VKA+SRVDKFSKSDIIVSPSILSANFA
Sbjct: 260 ING-FCLH-KTSL--SHPRSLTFSRKKISTTVKATSRVDKFSKSDIIVSPSILSANFA 421
>BE611450 similar to GP|6007803|gb|A D-ribulose-5-phosphate 3-epimerase
{Oryza sativa}, partial (34%)
Length = 340
Score = 57.8 bits (138), Expect = 5e-09
Identities = 38/116 (32%), Positives = 60/116 (50%), Gaps = 3/116 (2%)
Frame = -1
Query: 149 VKSLGAKAGVVLNPGTPLSAIEYVLDV---VDLVLIMSVNPGFGGQSFIESQVKKISDLR 205
+KS + V TPL + V+ D+V++ +V GFG Q F+ K+
Sbjct: 337 IKSHSKTSCVASKRATPLKKVILVVKAGIRTDMVIVNTVEQGFGEQKFM*EMTDKV---- 170
Query: 206 RLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTS 261
R+ ++K + IEVDGG+ P+ AGAN +V GS+VFGA + + I ++ S
Sbjct: 169 RIRSKKHPSFDIEVDGGLGPSTINVTASAGANCIVDGSSVFGALEPVQVIS*LRNS 2
>BU965495
Length = 431
Score = 41.2 bits (95), Expect = 5e-04
Identities = 15/35 (42%), Positives = 26/35 (73%)
Frame = +1
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDG 82
++PS+LS++FA L + + + G DW+H+D+MDG
Sbjct: 34 IAPSMLSSDFANLASEAQRMLHFGADWLHMDIMDG 138
>BG155087
Length = 184
Score = 35.8 bits (81), Expect = 0.021
Identities = 17/39 (43%), Positives = 25/39 (63%)
Frame = +2
Query: 217 IEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAI 255
++VDGG+ P+ A AN +VAGS+V GA + A+ I
Sbjct: 29 LQVDGGLWPSTIDVAASARANCIVAGSSVIGAPEPAQVI 145
>TC216169 weakly similar to UP|Q6JJ29 (Q6JJ29) Prephenate dehydratase,
partial (64%)
Length = 1640
Score = 29.3 bits (64), Expect = 1.9
Identities = 21/77 (27%), Positives = 36/77 (46%), Gaps = 4/77 (5%)
Frame = -3
Query: 120 PDFIKAGADIVSIH----CEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDV 175
PD G +S H ++S+ LH TV+Q+ + A + +PG PL I +
Sbjct: 639 PDVFSIGRITLSAHHISTALKASSKSLHETVSQLG*ALSAASSLYDPGIPLYDIRTLA*S 460
Query: 176 VDLVLIMSVNPGFGGQS 192
+ I ++ GFG ++
Sbjct: 459 SEAATISDMDNGFGRET 409
>TC206243 similar to PIR|B86314|B86314 F2H15.12 protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (95%)
Length = 1572
Score = 28.9 bits (63), Expect = 2.5
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Frame = +2
Query: 131 SIHCEQSSTIH-LHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIM 182
+ H E S + L R ++ K GAK VV G+PL +V D+ D V+ M
Sbjct: 791 NFHPENSHVLPALMRRFHEAKVNGAKEVVVWGTGSPLREFLHVDDLADAVVFM 949
>BM086666 similar to SP|O82660|H136_ Photosystem II stability/assembly factor
HCF136 chloroplast precursor. [Mouse-ear cress],
partial (11%)
Length = 428
Score = 28.9 bits (63), Expect = 2.5
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 3/46 (6%)
Frame = +1
Query: 18 PRSLTFSRRKISTIVKASSRVDKFSKSD---IIVSPSILSANFAKL 60
P + TFSR + S IVKASS D FS+S I + LS + +L
Sbjct: 133 PSTRTFSRNR-SLIVKASSSEDDFSRSSRRRFIAETAALSVSLPQL 267
>TC206245 similar to UP|O49213 (O49213) GDP-4-keto-6-deoxy-D-mannose-3,
5-epimerase-4-reductase (GER1); 21556-22494, partial
(57%)
Length = 582
Score = 28.9 bits (63), Expect = 2.5
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Frame = +3
Query: 131 SIHCEQSSTIH-LHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIM 182
+ H E S + L R ++ K GAK VV G+PL +V D+ D V+ M
Sbjct: 126 NFHPENSHVLPALMRRFHEAKVNGAKEVVVWGTGSPLREFLHVDDLADAVVFM 284
>TC221610
Length = 519
Score = 28.5 bits (62), Expect = 3.3
Identities = 20/56 (35%), Positives = 31/56 (54%)
Frame = -3
Query: 15 NNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELA 70
N+ SL RK S + A+ RV F+ S + VSPSI+ +NF + ++ V +A
Sbjct: 337 NSPYSSLALKMRK*S--IPATIRVLYFNPSTVRVSPSIIFSNFFFVPNRLSKVAMA 176
>BG405288 homologue to GP|20339362|gb| ribulose-5-phosphate-3-epimerase
{Pisum sativum}, partial (4%)
Length = 413
Score = 28.1 bits (61), Expect = 4.3
Identities = 19/42 (45%), Positives = 21/42 (49%)
Frame = +1
Query: 163 GTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDL 204
GTPLSAIEY+LD +L F F K ISDL
Sbjct: 4 GTPLSAIEYILDGEPFLL------PFACLLFFSLYNKNISDL 111
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,711,626
Number of Sequences: 63676
Number of extensions: 110089
Number of successful extensions: 492
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 482
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 484
length of query: 268
length of database: 12,639,632
effective HSP length: 96
effective length of query: 172
effective length of database: 6,526,736
effective search space: 1122598592
effective search space used: 1122598592
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0093a.1


