

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0090b.2
(120 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
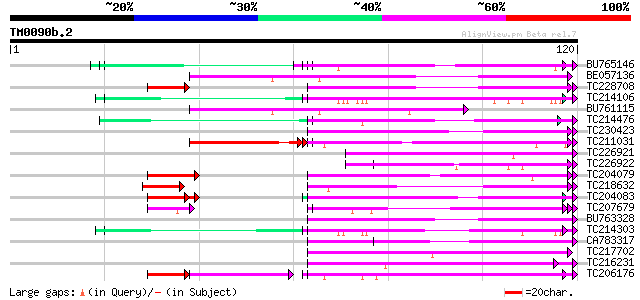
Score E
Sequences producing significant alignments: (bits) Value
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 60 1e-10
BE057136 similar to PIR|JQ1060|JQ10 glycine-rich protein 1 - Ara... 57 9e-10
TC228708 similar to UP|GRP2_PHAVU (P10496) Glycine-rich cell wal... 49 5e-09
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 54 1e-08
BU761115 similar to PIR|S17732|KNMU glycine-rich cell wall prote... 54 1e-08
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 53 2e-08
TC230423 similar to UP|Q41187 (Q41187) Glycine-rich protein (Fra... 52 4e-08
TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 45 4e-08
TC226921 similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein p... 52 5e-08
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 51 7e-08
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 50 1e-07
TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wal... 47 2e-07
TC204083 similar to UP|O48567 (O48567) Glycine-rich RNA-binding ... 43 3e-07
TC207679 similar to UP|Q7XJI7 (Q7XJI7) Glycine-rich protein TomR... 47 5e-07
BU763328 similar to SP|P10496|GRP2_ Glycine-rich cell wall struc... 48 6e-07
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 48 6e-07
CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 48 7e-07
TC217702 similar to GB|AAL06971.1|15810091|AY056083 AT4g30260/F9... 48 7e-07
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 47 2e-06
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 45 2e-06
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 60.1 bits (144), Expect = 1e-10
Identities = 29/58 (50%), Positives = 33/58 (56%)
Frame = +1
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GGGG GGG GG GGG + G R R+ + GGG G GGG GGG GGGG
Sbjct: 154 EGGGGGGGGGGGGGGGGGGGVF*GGERX----REGGERGGGGGGGGGGGGGGGXGGGG 315
Score = 58.9 bits (141), Expect = 3e-10
Identities = 37/101 (36%), Positives = 40/101 (38%)
Frame = +1
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGG 79
EGGG GGGG +GGGG GGG GG GG
Sbjct: 100 EGGGGGGGGGGGGGGGGG-------------------------EGGGGGGGGGGGGGGGG 204
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G + G R + GGG G GGG GGGGGGGG
Sbjct: 205 GGGVF*GGERXREGGERGGGGGGGGGGGGGGGXGGGGGGGG 327
Score = 58.2 bits (139), Expect = 5e-10
Identities = 29/57 (50%), Positives = 29/57 (50%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G R GGG G GGG GGGGGGGG
Sbjct: 26 GGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGG 196
Score = 58.2 bits (139), Expect = 5e-10
Identities = 29/57 (50%), Positives = 29/57 (50%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G R GGG G GGG GGGGGGGG
Sbjct: 29 GGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGG 199
Score = 57.4 bits (137), Expect = 9e-10
Identities = 28/57 (49%), Positives = 29/57 (50%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G + GGG G GGG GGGGGGGG
Sbjct: 21 GGGGGGGGGGGGGGGGGGGGGGG------------EGGGGGGGGGGGGGGGGGGGGG 155
Score = 57.0 bits (136), Expect = 1e-09
Identities = 36/100 (36%), Positives = 37/100 (37%)
Frame = +3
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG GGGG+ R GGGG GGG GG GGG
Sbjct: 189 GGGGGGGGGFLGGRE--------------------------XXGGGGEGGGGGGGGGGGG 290
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GGG G GGG GGGGGGGG
Sbjct: 291 GGGXGG-------------GGGGRGEGGGGGGGGGGGGGG 371
Score = 57.0 bits (136), Expect = 1e-09
Identities = 28/57 (49%), Positives = 29/57 (50%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG +GGG GG GGG G GGG G GGG GGGGGGGG
Sbjct: 75 GGGGGEGGGGGGGGGGGGGGGGG-------------GGGGRGGGGGGGGGGGGGGGG 206
Score = 57.0 bits (136), Expect = 1e-09
Identities = 27/56 (48%), Positives = 30/56 (53%)
Frame = +2
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG ++ R R + GGG G GGG GGGGGG G
Sbjct: 158 GGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEG 325
Score = 56.6 bits (135), Expect = 2e-09
Identities = 30/62 (48%), Positives = 32/62 (51%), Gaps = 2/62 (3%)
Frame = +1
Query: 61 VVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG--GGGG 118
V GGGG GGG GG GGG GR + GGG G +GGG GG GGGG
Sbjct: 13 VCPGGGGGGGGGGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGGGG 192
Query: 119 GG 120
GG
Sbjct: 193 GG 198
Score = 56.2 bits (134), Expect = 2e-09
Identities = 28/57 (49%), Positives = 31/57 (54%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG G ++R + GG G GGG GGGGGGGG
Sbjct: 143 GGGGKGGGGGGGGGGGGGGGGGGG---FFRGERXXGRGGRGGGGGGGGGGGGGGGGG 304
Score = 55.8 bits (133), Expect = 3e-09
Identities = 36/100 (36%), Positives = 38/100 (38%)
Frame = +2
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG GGGG ++R R G GG GGG GG GGG
Sbjct: 182 GGGGGGGGGGGFFRGERXX------------------------GRGGRGGGGGGGGGGGG 289
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GGG G GGG GGGGGGGG
Sbjct: 290 GGGGGG-------------GGGGEGGGGGGGGGGGGGGGG 370
Score = 54.3 bits (129), Expect = 8e-09
Identities = 36/103 (34%), Positives = 37/103 (34%)
Frame = +1
Query: 18 VVEGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSS 77
V GGG GGGG GGGG GGG GG
Sbjct: 13 VCPGGGGGGGGG-----------------------------GGGGGGGGGGGGGGRGGEG 105
Query: 78 GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G GGG G +GGG GGGGGGGG
Sbjct: 106 GGGGGGGGG--------------GGGGGGGEGGGGGGGGGGGG 192
Score = 53.5 bits (127), Expect = 1e-08
Identities = 27/57 (47%), Positives = 27/57 (47%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G GGG GGGGGGGG
Sbjct: 262 GGGGGGGGGGGGGXGGG--------------------GGGGGRGGGGGGGGGGGGGG 372
Score = 53.1 bits (126), Expect = 2e-08
Identities = 27/57 (47%), Positives = 28/57 (48%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG G GGG GG GGG G GGG G + GGG GGGGGGGG
Sbjct: 3 GGRGLPGGGGGGGGGGGGGGGGG--------------GGGGGGEGGGGGGGGGGGGG 131
Score = 52.0 bits (123), Expect = 4e-08
Identities = 34/101 (33%), Positives = 36/101 (34%)
Frame = +3
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGG 79
EGGG GGGG GGGG GGG G GG
Sbjct: 90 EGGGGGGGGGG--------------------------------GGGGGGGGGGGRGGGGG 173
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G G + + GG G GGG GGGGGGGG
Sbjct: 174 GGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGGGGGGGG 296
Score = 50.8 bits (120), Expect = 9e-08
Identities = 26/57 (45%), Positives = 26/57 (45%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G GGG GGGGGG G
Sbjct: 265 GGGGGGGGGGGGXGGGGG-------------------GGGRGGGGGGGGGGGGGGEG 378
Score = 50.4 bits (119), Expect = 1e-07
Identities = 28/62 (45%), Positives = 28/62 (45%), Gaps = 5/62 (8%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG-----GGGG 118
GGGG GGG GG GGG R GGG G GGG GG GGGG
Sbjct: 274 GGGGGGGGGXGGGGGGGGR------------------GGGGGGGGGGGGGGEGREEGGGG 399
Query: 119 GG 120
GG
Sbjct: 400 GG 405
Score = 49.3 bits (116), Expect = 3e-07
Identities = 27/58 (46%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Frame = +1
Query: 65 GGGN--DGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG G GGG G + GGG GGGGGGGG
Sbjct: 1 GGGEVCPGGGGGGGGGGGGGGGGGG-----------GGGGGRGGEGGGGGGGGGGGGG 141
Score = 46.2 bits (108), Expect = 2e-06
Identities = 26/59 (44%), Positives = 26/59 (44%), Gaps = 4/59 (6%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG----GGGG 118
GGGG GGG GG GGG G GGG G GGG GG GGGG
Sbjct: 266 GGGGGGGGGGGGGGGGGGEGGGG------------GGGGGGGGGGGGGRGGRREEGGGG 406
Score = 24.3 bits (51), Expect = 8.7
Identities = 19/63 (30%), Positives = 19/63 (30%), Gaps = 2/63 (3%)
Frame = +2
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGG--SS 77
EGGG GGG GGGG GGG GG
Sbjct: 320 EGGGGGGGG-----------------------------------GGGGGGGGGRGGRREE 394
Query: 78 GGG 80
GGG
Sbjct: 395 GGG 403
>BE057136 similar to PIR|JQ1060|JQ10 glycine-rich protein 1 - Arabidopsis
thaliana (fragment), partial (19%)
Length = 420
Score = 57.4 bits (137), Expect = 9e-10
Identities = 38/83 (45%), Positives = 45/83 (53%), Gaps = 2/83 (2%)
Frame = -3
Query: 39 SVVVVAVTVVVIVVEA-VVVEAVVVDG-GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQ 96
SV+ VA +V+ V E + E +VDG GGG GGG+GG GGGA A G
Sbjct: 217 SVLAVAFVLVIGVAECRKIKENELVDGFGGGGLGGGAGGGFGGGAGAGIG---------- 68
Query: 97 WCDDGGGSGSDDGGGSGGGGGGG 119
GGG+G GGG GGG GGG
Sbjct: 67 ---GGGGAGGGAGGGFGGGKGGG 8
Score = 25.4 bits (54), Expect = 3.9
Identities = 8/10 (80%), Positives = 8/10 (80%), Gaps = 1/10 (10%)
Frame = -2
Query: 29 GW-CWWRRWR 37
GW CWWR WR
Sbjct: 53 GWGCWWRFWR 24
Score = 24.3 bits (51), Expect = 8.7
Identities = 8/12 (66%), Positives = 8/12 (66%)
Frame = -2
Query: 28 GGWCWWRRWRRS 39
G WC WR W RS
Sbjct: 119 GWWCRWRFWWRS 84
>TC228708 similar to UP|GRP2_PHAVU (P10496) Glycine-rich cell wall structural
protein 1.8 precursor (GRP 1.8), partial (15%)
Length = 494
Score = 48.5 bits (114), Expect(2) = 5e-09
Identities = 27/57 (47%), Positives = 28/57 (48%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG DGG GG GGGA A G G G+G GGG GGGGG GG
Sbjct: 66 GGGYGDGGAHGGGYGGGAGAGGG-------------GGYGAGGAHGGGFGGGGGSGG 197
Score = 43.1 bits (100), Expect = 2e-05
Identities = 24/56 (42%), Positives = 25/56 (43%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G G GGSGG SGGG DGG G GGG+G GGGGG
Sbjct: 24 GXGYGGAGGSGGGSGGGY-----------------GDGGAHGGGYGGGAGAGGGGG 140
Score = 32.0 bits (71), Expect = 0.042
Identities = 19/55 (34%), Positives = 21/55 (37%)
Frame = +3
Query: 66 GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG G G GG+ G G GG G GG+ GGG GGG
Sbjct: 12 GGEHGXGYGGAGGSG--------------------GGSGGGYGDGGAHGGGYGGG 116
Score = 26.2 bits (56), Expect = 2.3
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = +3
Query: 100 DGGGSGSDDGGGSGGGGGGGG 120
+ GG GG+GG GGG G
Sbjct: 6 EAGGEHGXGYGGAGGSGGGSG 68
Score = 26.2 bits (56), Expect(2) = 5e-09
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +2
Query: 30 WCWWRRWRR 38
W WWR WRR
Sbjct: 47 WKWWREWRR 73
Score = 25.8 bits (55), Expect = 3.0
Identities = 9/20 (45%), Positives = 11/20 (55%), Gaps = 1/20 (5%)
Frame = +2
Query: 79 GGARAWWGRRRQWWR-WRQW 97
G R W R WW+ WR+W
Sbjct: 8 GRWRTWHXLWRSWWKWWREW 67
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 53.5 bits (127), Expect = 1e-08
Identities = 27/57 (47%), Positives = 29/57 (50%)
Frame = -2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG N GGG GG GGG G Q+ + GGG G GGG GGGG GG
Sbjct: 179 GGGENTGGGGGGGDGGGGEGGGGDLTQYVGEGGGGEGGGGGGE*TGGGGEGGGGDGG 9
Score = 48.9 bits (115), Expect = 3e-07
Identities = 34/98 (34%), Positives = 35/98 (35%)
Frame = -2
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGGD GGGW A DGGGG GG GGG
Sbjct: 524 GGGDGDGGGW*G--------------------------AGCGDGGGGECSSHGGGDGGGG 423
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
+ G GGG GS GGG GGGGGG
Sbjct: 422 GGSIQG-------------GGGGGGSIQGGGDGGGGGG 348
Score = 47.0 bits (110), Expect = 1e-06
Identities = 25/57 (43%), Positives = 28/57 (48%)
Frame = -2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG G GGG G + + C GGG + GGG G GGGG G
Sbjct: 281 GGGGEGGGGGGECGGGGGGECCGGGGEL**Y---CTGGGGENTGGGGGGGDGGGGEG 120
Score = 47.0 bits (110), Expect = 1e-06
Identities = 32/70 (45%), Positives = 32/70 (45%), Gaps = 12/70 (17%)
Frame = -2
Query: 63 DGGGGN---DGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS------GSDDGGGSG 113
DGGGG GGG G GGG W G C DGGG G D GGG G
Sbjct: 560 DGGGGELY*TGGGGGDGDGGG---W*GAG---------CGDGGGGECSSHGGGDGGGGGG 417
Query: 114 ---GGGGGGG 120
GGGGGGG
Sbjct: 416 SIQGGGGGGG 387
Score = 46.6 bits (109), Expect = 2e-06
Identities = 30/70 (42%), Positives = 31/70 (43%), Gaps = 13/70 (18%)
Frame = -2
Query: 64 GGGGNDGGG-----SGGSSGGGARAWWGRRRQWWRWRQWCDDGGGS-----GSDDGGGSG 113
GGGG+ GGG GG G W G C DGGG G DGGG G
Sbjct: 572 GGGGDGGGGELY*TGGGGGDGDGGGW*G---------AGCGDGGGGECSSHGGGDGGGGG 420
Query: 114 G---GGGGGG 120
G GGGGGG
Sbjct: 419 GSIQGGGGGG 390
Score = 45.8 bits (107), Expect = 3e-06
Identities = 37/116 (31%), Positives = 41/116 (34%), Gaps = 14/116 (12%)
Frame = -2
Query: 19 VEGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGND------GGG 72
++GGGD GGGG E GGGG D GGG
Sbjct: 383 IQGGGDGGGGGG----------------------E*TGGGGE*TGGGGGGDK*G*TGGGG 270
Query: 73 SGGSSGGGARAWWGRRRQWWRWRQWCDDG--------GGSGSDDGGGSGGGGGGGG 120
GG GG G + C G GG G + GGG GGG GGGG
Sbjct: 269 EGGGGGGECGGGGG--------GECCGGGGEL**YCTGGGGENTGGGGGGGDGGGG 126
Score = 45.1 bits (105), Expect = 5e-06
Identities = 30/70 (42%), Positives = 31/70 (43%), Gaps = 13/70 (18%)
Frame = -2
Query: 64 GGGGNDGGGSG------GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGG-- 115
GGGG DG G G G GGG + G D GGG GS GGG GGG
Sbjct: 530 GGGGGDGDGGGW*GAGCGDGGGGECSSHGGG----------DGGGGGGSIQGGGGGGGSI 381
Query: 116 -----GGGGG 120
GGGGG
Sbjct: 380 QGGGDGGGGG 351
Score = 41.6 bits (96), Expect = 5e-05
Identities = 26/64 (40%), Positives = 28/64 (43%), Gaps = 6/64 (9%)
Frame = -2
Query: 63 DGGGGNDGGGS----GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSD--DGGGSGGGG 116
DGGGG GGG G GGG +GGG G + GGG GGGG
Sbjct: 140 DGGGGEGGGGDLTQYVGEGGGG-------------------EGGGGGGE*TGGGGEGGGG 18
Query: 117 GGGG 120
GGG
Sbjct: 17 DGGG 6
Score = 40.8 bits (94), Expect = 9e-05
Identities = 33/103 (32%), Positives = 35/103 (33%), Gaps = 3/103 (2%)
Frame = -2
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDG---GGSGGSS 77
GGGD GGGG + GGGG G GG G
Sbjct: 446 GGGDGGGGGGS------------------------------IQGGGGGGGSIQGGGDGGG 357
Query: 78 GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G GGG G D G +GGGG GGG
Sbjct: 356 GGGE*TGGGGE----------*TGGGGGGDK*G*TGGGGEGGG 258
Score = 37.0 bits (84), Expect = 0.001
Identities = 28/70 (40%), Positives = 30/70 (42%), Gaps = 12/70 (17%)
Frame = -2
Query: 63 DGGGGND----GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGG------SGSDDGGGS 112
DGGGG GGG GGS GG G + GGG +G GG
Sbjct: 437 DGGGGGGSIQGGGGGGGSIQGGGDGGGGGGE*TGGGGE*TGGGGGGDK*G*TGGGGEGGG 258
Query: 113 GGG--GGGGG 120
GGG GGGGG
Sbjct: 257 GGGECGGGGG 228
Score = 32.3 bits (72), Expect = 0.032
Identities = 26/92 (28%), Positives = 30/92 (32%)
Frame = -2
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGG 79
+GGG GGGG + V +GGGG GGG G +GG
Sbjct: 140 DGGGGEGGGG--------------------------DLTQYVGEGGGGEGGGGGGE*TGG 39
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGG 111
G G G G D GGG
Sbjct: 38 G--------------------GEGGGGDGGGG 3
Score = 28.5 bits (62), Expect = 0.46
Identities = 22/66 (33%), Positives = 26/66 (39%), Gaps = 4/66 (6%)
Frame = -2
Query: 58 EAVVVDGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGG----SG 113
E + G G + G GG+ GGG G G G D GGG +G
Sbjct: 668 ETPMTGGKGPSYTGVGGGNDGGGGEL***T-------------GVGGGGDGGGGELY*TG 528
Query: 114 GGGGGG 119
GGGG G
Sbjct: 527 GGGGDG 510
Score = 28.1 bits (61), Expect = 0.60
Identities = 14/28 (50%), Positives = 15/28 (53%), Gaps = 8/28 (28%)
Frame = -2
Query: 101 GGGSGSDDGGGS--------GGGGGGGG 120
G G G+D GGG GGG GGGG
Sbjct: 629 GVGGGNDGGGGEL***TGVGGGGDGGGG 546
Score = 28.1 bits (61), Expect = 0.60
Identities = 10/14 (71%), Positives = 10/14 (71%), Gaps = 2/14 (14%)
Frame = -1
Query: 84 WWG--RRRQWWRWR 95
WW RRR WWRWR
Sbjct: 42 WWR*RRRR*WWRWR 1
>BU761115 similar to PIR|S17732|KNMU glycine-rich cell wall protein precursor
- Arabidopsis thaliana, partial (11%)
Length = 274
Score = 53.5 bits (127), Expect = 1e-08
Identities = 31/65 (47%), Positives = 39/65 (59%), Gaps = 6/65 (9%)
Frame = +1
Query: 39 SVVVVAVTVVVIVVEA-VVVEAVVVDG-GGGNDGGGSGGSSGGGARA----WWGRRRQWW 92
SV+ VA +V+ V E + E +VDG GGG GGG+GG GGGA A WW R +W
Sbjct: 70 SVLAVAFVLVIGVAECRKIKENELVDGFGGGGLGGGAGGGFGGGAGAGIGMWWWCRWGFW 249
Query: 93 RWRQW 97
RW +W
Sbjct: 250 RW*RW 264
Score = 33.1 bits (74), Expect = 0.019
Identities = 17/34 (50%), Positives = 19/34 (55%), Gaps = 4/34 (11%)
Frame = +3
Query: 28 GGWC----WWRRWRRSVVVVAVTVVVIVVEAVVV 57
G WC WWR W R+ VV V V V+ V VVV
Sbjct: 168 GWWCRWRFWWRSWCRNWYVVVVQVGVLEVVKVVV 269
Score = 32.7 bits (73), Expect = 0.024
Identities = 19/40 (47%), Positives = 22/40 (54%)
Frame = +2
Query: 41 VVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
V+ V VV+ VE +V E V GG GGG GG GGG
Sbjct: 149 VLEEVDWVVVPVEVLVEELVQELVCGGGAGGGFGGGKGGG 268
Score = 29.6 bits (65), Expect = 0.21
Identities = 12/19 (63%), Positives = 12/19 (63%)
Frame = +1
Query: 101 GGGSGSDDGGGSGGGGGGG 119
GGG G GGG GGG G G
Sbjct: 157 GGGLGGGAGGGFGGGAGAG 213
Score = 27.3 bits (59), Expect(2) = 0.006
Identities = 11/17 (64%), Positives = 12/17 (69%)
Frame = +2
Query: 101 GGGSGSDDGGGSGGGGG 117
GGG+G GGG GGG G
Sbjct: 224 GGGAGGGFGGGKGGGIG 274
Score = 26.6 bits (57), Expect(2) = 0.006
Identities = 9/15 (60%), Positives = 9/15 (60%)
Frame = +3
Query: 84 WWGRRRQWWRWRQWC 98
WW R R W WR WC
Sbjct: 171 WWCRWRFW--WRSWC 209
Score = 26.6 bits (57), Expect = 1.8
Identities = 13/20 (65%), Positives = 14/20 (70%)
Frame = +1
Query: 100 DGGGSGSDDGGGSGGGGGGG 119
DG G G GGG+GGG GGG
Sbjct: 145 DGFGGGGL-GGGAGGGFGGG 201
Score = 26.2 bits (56), Expect = 2.3
Identities = 9/16 (56%), Positives = 9/16 (56%)
Frame = +1
Query: 21 GGGDSGGGGWCWWRRW 36
GGG G G WW RW
Sbjct: 193 GGGAGAGIGMWWWCRW 240
Score = 25.4 bits (54), Expect = 3.9
Identities = 11/24 (45%), Positives = 11/24 (45%), Gaps = 7/24 (29%)
Frame = +1
Query: 21 GGGDSGGGG-------WCWWRRWR 37
GGG GG G WC W WR
Sbjct: 181 GGGFGGGAGAGIGMWWWCRWGFWR 252
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 53.1 bits (126), Expect = 2e-08
Identities = 27/57 (47%), Positives = 29/57 (50%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GGGSG GG GGGSG +GGGS GGGGGGG
Sbjct: 469 GGGGSQGGGSGSGGGGYG-------------------GGGSGGSEGGGSSGGGGGGG 356
Score = 52.0 bits (123), Expect = 4e-08
Identities = 26/57 (45%), Positives = 28/57 (48%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GG GG SG G + G GG GS+ GG SGGGGGGGG
Sbjct: 481 GGGSGGGGSQGGGSGSGGGGYGG--------------GGSGGSEGGGSSGGGGGGGG 353
Score = 51.2 bits (121), Expect = 7e-08
Identities = 27/57 (47%), Positives = 28/57 (48%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGGSGGS GGG+ GG G GGG GGG GGGG
Sbjct: 433 GGGGYGGGGSGGSEGGGS-------------------SGGGGGGGGGGGGGGYGGGG 320
Score = 46.2 bits (108), Expect = 2e-06
Identities = 27/60 (45%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Frame = -3
Query: 64 GGGGNDGGGS---GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG+ GGG GGS GGG A G GG +G +G GS GGG GGG
Sbjct: 616 GGGGSKGGGGYGGGGSQGGGGSAGGGGS*G--------GSGGSAGGGEGSGSQGGGSGGG 461
Score = 43.9 bits (102), Expect = 1e-05
Identities = 28/57 (49%), Positives = 28/57 (49%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGGS G SGG A G Q GGGSG GGGS GGG G G
Sbjct: 562 GGGSAGGGGS*GGSGGSAGGGEGSGSQ----------GGGSG---GGGSQGGGSGSG 431
Score = 43.1 bits (100), Expect = 2e-05
Identities = 32/98 (32%), Positives = 33/98 (33%)
Frame = -3
Query: 20 EGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGG 79
EGGG SGGGG GGGG G G GG GG
Sbjct: 394 EGGGSSGGGGG-------------------------------GGGGGGGGGYGGGGDKGG 308
Query: 80 GARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGG 117
G G + GG G DGG GGGGG
Sbjct: 307 GIGGDKG---------DYHHGKGGKGDKDGGDKGGGGG 221
Score = 39.3 bits (90), Expect = 3e-04
Identities = 22/56 (39%), Positives = 22/56 (39%)
Frame = -3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GG G GGG GG GS GGGS GGGG G
Sbjct: 634 GGGGSAGGGGSKGGGGY--------------------GGGGSQGGGGSAGGGGS*G 527
Score = 31.6 bits (70), Expect = 0.055
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = -3
Query: 100 DGGGSGSDDGGGSGGGGGGGG 120
+GGG + GG GGGG GGG
Sbjct: 637 EGGGGSAGGGGSKGGGGYGGG 575
Score = 26.2 bits (56), Expect = 2.3
Identities = 12/17 (70%), Positives = 13/17 (75%), Gaps = 2/17 (11%)
Frame = -3
Query: 106 SDDGGGSGGGGG--GGG 120
S+ GGGS GGGG GGG
Sbjct: 640 SEGGGGSAGGGGSKGGG 590
>TC230423 similar to UP|Q41187 (Q41187) Glycine-rich protein (Fragment),
partial (40%)
Length = 522
Score = 52.0 bits (123), Expect = 4e-08
Identities = 28/57 (49%), Positives = 31/57 (54%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG +GGGA +G GGG G GGG+GGG GGGG
Sbjct: 107 GGGGGAGGGFGGGAGGGAGGGFGGGAGGGA-------GGGFGGGAGGGAGGGFGGGG 256
Score = 46.2 bits (108), Expect = 2e-06
Identities = 25/57 (43%), Positives = 27/57 (46%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG+GG GGG GGG G GGG+GGGGG GG
Sbjct: 17 GGGIGGGGGAGGGFGGG-------------------HGGGVGGGIGGGAGGGGGAGG 130
Score = 46.2 bits (108), Expect = 2e-06
Identities = 25/56 (44%), Positives = 25/56 (44%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GGG G G G G GGG GGG GGG
Sbjct: 29 GGGGGAGGGFGGGHGGGVGGGIG-------------GGAGGGGGAGGGFGGGAGGG 157
Score = 31.2 bits (69), Expect = 0.071
Identities = 13/20 (65%), Positives = 13/20 (65%)
Frame = +2
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
GGG G GGG G GGG GG
Sbjct: 5 GGGVGGGIGGGGGAGGGFGG 64
Score = 28.9 bits (63), Expect = 0.35
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +2
Query: 101 GGGSGSDDGGGSGGGGGGG 119
G G G GGG+GGG GGG
Sbjct: 11 GVGGGIGGGGGAGGGFGGG 67
Score = 26.6 bits (57), Expect = 1.8
Identities = 19/73 (26%), Positives = 21/73 (28%), Gaps = 7/73 (9%)
Frame = +1
Query: 32 WWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGGARAW-WGRRRQ 90
WWRRW R + G + G R W W R R
Sbjct: 34 WWRRWWR------------------LRRWTWWWSRGRNWWRCRWRRRSGRRLWRWSRWRS 159
Query: 91 WW------RWRQW 97
WW RWR W
Sbjct: 160WWWLWRWCRWRSW 198
Score = 25.0 bits (53), Expect = 5.1
Identities = 8/14 (57%), Positives = 9/14 (64%)
Frame = +1
Query: 84 WWGRRRQWWRWRQW 97
W RRR W WR+W
Sbjct: 7 WRCRRRNRWWWRRW 48
>TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (11%)
Length = 678
Score = 45.4 bits (106), Expect(2) = 4e-08
Identities = 24/57 (42%), Positives = 28/57 (49%)
Frame = -3
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
+G GG GGG GG GG G + + GGG G + GGG GG GGGG
Sbjct: 358 EGDGGKGGGGDGGKGGGVDSGIGGGKGGGG---DGGNGGGGDGGNGGGGDGGNGGGG 197
Score = 45.1 bits (105), Expect = 5e-06
Identities = 27/61 (44%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Frame = -3
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGG---GSGGGGGGG 119
+GG G GG SGG+ GGGA G ++ GGG G GG G GGG GGG
Sbjct: 445 NGGSGGGGGASGGNGGGGASG--GGDGRFGGEGDGGKGGGGDGGKGGGVDSGIGGGKGGG 272
Query: 120 G 120
G
Sbjct: 271 G 269
Score = 45.1 bits (105), Expect(2) = 1e-07
Identities = 25/59 (42%), Positives = 26/59 (43%), Gaps = 3/59 (5%)
Frame = -3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG---GGGG 120
GGG GGG GG+ GGG DGG G DGG GGG GGGG
Sbjct: 292 GGGKGGGGDGGNGGGG-------------------DGGNGGGGDGGNGGGGASGIGGGG 173
Score = 43.5 bits (101), Expect = 1e-05
Identities = 24/57 (42%), Positives = 26/57 (45%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GGN GGG GG+ GGG +GGG S GGG GGGGG
Sbjct: 271 GDGGNGGGGDGGNGGGGDGG----------------NGGGGASGIGGGGHSLGGGGG 149
Score = 42.7 bits (99), Expect = 2e-05
Identities = 25/59 (42%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Frame = -3
Query: 62 VDGG-GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
VD G GG GGG G +GGG G GGG G + GGG+ G GGGG
Sbjct: 307 VDSGIGGGKGGGGDGGNGGGGDGGNG--------------GGGDGGNGGGGASGIGGGG 173
Score = 31.2 bits (69), Expect = 0.071
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = -3
Query: 102 GGSGSDDGGGSGGGGGGGG 120
G GS GGG+ GG GGGG
Sbjct: 448 GNGGSGGGGGASGGNGGGG 392
Score = 30.0 bits (66), Expect = 0.16
Identities = 18/50 (36%), Positives = 21/50 (42%)
Frame = -3
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS 112
+GGGG+ G G GG+SG G GGG GGGS
Sbjct: 235 NGGGGDGGNGGGGASGIG--------------------GGGHSLGGGGGS 146
Score = 30.0 bits (66), Expect = 0.16
Identities = 19/48 (39%), Positives = 20/48 (41%)
Frame = -3
Query: 72 GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GS GGG R G + G G GG SGG GGGG
Sbjct: 508 GRFGSRGGGTRYSPG---------PFGGIGNGGSGGGGGASGGNGGGG 392
Score = 26.2 bits (56), Expect(2) = 4e-08
Identities = 16/24 (66%), Positives = 17/24 (70%)
Frame = -1
Query: 39 SVVVVAVTVVVIVVEAVVVEAVVV 62
S + VAV VVV VV VVVE VVV
Sbjct: 450 SAMGVAVVVVVQVVVMVVVEPVVV 379
Score = 25.0 bits (53), Expect(2) = 1e-07
Identities = 15/25 (60%), Positives = 19/25 (76%)
Frame = -1
Query: 39 SVVVVAVTVVVIVVEAVVVEAVVVD 63
+VVVV VV++VVE VVV V+VD
Sbjct: 435 AVVVVVQVVVMVVVEPVVV--VMVD 367
Score = 25.0 bits (53), Expect = 5.1
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -3
Query: 64 GGGGNDGGGSGGSS 77
GGGG+ GG GGSS
Sbjct: 184 GGGGHSLGGGGGSS 143
>TC226921 similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein precursor,
partial (5%)
Length = 641
Score = 51.6 bits (122), Expect = 5e-08
Identities = 26/54 (48%), Positives = 29/54 (53%), Gaps = 5/54 (9%)
Frame = -2
Query: 72 GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSG-----SDDGGGSGGGGGGGG 120
G G + G R W RRR+ WRW W GGSG + GGG G GGGGGG
Sbjct: 208 GRGRRASPGRRRRWCRRRRVWRWWWWEGGEGGSGGGALRAPGGGGGGWGGGGGG 47
Score = 29.6 bits (65), Expect = 0.21
Identities = 19/58 (32%), Positives = 22/58 (37%)
Frame = -2
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
+GG G GGG+ + GGG GGG GGGGGG G
Sbjct: 130 EGGEGGSGGGALRAPGGG----------------------------GGGWGGGGGGRG 41
Score = 25.0 bits (53), Expect = 5.1
Identities = 13/26 (50%), Positives = 14/26 (53%)
Frame = -2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRR 89
GGGG GGG GG R GRR+
Sbjct: 82 GGGGGWGGGGGGRGPSRPR---GRRK 14
Score = 24.6 bits (52), Expect = 6.7
Identities = 11/29 (37%), Positives = 15/29 (50%)
Frame = -3
Query: 54 AVVVEAVVVDGGGGNDGGGSGGSSGGGAR 82
AV + ++ GGG+ G G GGG R
Sbjct: 213 AVAEDGELLQVGGGDGVAGGGFGDGGGGR 127
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 51.2 bits (121), Expect = 7e-08
Identities = 27/53 (50%), Positives = 28/53 (51%), Gaps = 4/53 (7%)
Frame = -2
Query: 72 GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSD----DGGGSGGGGGGGG 120
G G + G R W RRR WRW W GGGSG GGG GG GGGGG
Sbjct: 241 GQGWRASRGRRRQWRRRRLVWRW-WWEGGGGGSGGGARRAPGGGDGGWGGGGG 86
Score = 47.4 bits (111), Expect = 1e-06
Identities = 27/55 (49%), Positives = 28/55 (50%), Gaps = 13/55 (23%)
Frame = -2
Query: 78 GGGARAWWGRRRQWWR----WRQWCDDGGGS---------GSDDGGGSGGGGGGG 119
G G RA GRRRQW R WR W + GGG G DGG GGGGG G
Sbjct: 241 GQGWRASRGRRRQWRRRRLVWRWWWEGGGGGSGGGARRAPGGGDGGWGGGGGGRG 77
Score = 25.4 bits (54), Expect = 3.9
Identities = 14/34 (41%), Positives = 16/34 (46%), Gaps = 8/34 (23%)
Frame = -2
Query: 63 DGGGGNDGGGS--------GGSSGGGARAWWGRR 88
+GGGG GGG+ GG GGG RR
Sbjct: 166 EGGGGGSGGGARRAPGGGDGGWGGGGGGRGPSRR 65
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 50.1 bits (118), Expect = 1e-07
Identities = 27/56 (48%), Positives = 29/56 (51%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGGG GGG GG GG + G R + GGG G GGG GGGGGGG
Sbjct: 328 GGGGGGGGGGGGYGGGRGGGYGGGGR--GGGGGYNRSGGGGGYGGGGGYGGGGGGG 489
Score = 48.5 bits (114), Expect = 4e-07
Identities = 28/58 (48%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG-GGSGGGGGGGG 120
GGGG GGG GG GGG R G + GGG G G GG GGGG GGG
Sbjct: 349 GGGGGYGGGRGGGYGGGGRGGGGGYNR-------SGGGGGYGGGGGYGGGGGGGYGGG 501
Score = 43.1 bits (100), Expect(2) = 7e-07
Identities = 25/57 (43%), Positives = 25/57 (43%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG G GGG GR D G G G D G GG GG GG
Sbjct: 433 GGGGGYGGGGGYGGGGGGGYGGGR-----------DRGYGGGGDRGYSRGGDGGDGG 570
Score = 33.9 bits (76), Expect = 0.011
Identities = 14/20 (70%), Positives = 14/20 (70%)
Frame = +1
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
GGG G GGG GGG GGGG
Sbjct: 343 GGGGGGGYGGGRGGGYGGGG 402
Score = 32.0 bits (71), Expect = 0.042
Identities = 13/20 (65%), Positives = 13/20 (65%)
Frame = +1
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
GGG G GGG GGG GGG
Sbjct: 328 GGGGGGGGGGGGYGGGRGGG 387
Score = 29.3 bits (64), Expect = 0.27
Identities = 14/22 (63%), Positives = 14/22 (63%), Gaps = 2/22 (9%)
Frame = +1
Query: 101 GGGSGSDDGGGSGG--GGGGGG 120
GGG G GGG GG GGG GG
Sbjct: 331 GGGGGGGGGGGYGGGRGGGYGG 396
Score = 28.1 bits (61), Expect = 0.60
Identities = 11/14 (78%), Positives = 11/14 (78%)
Frame = +1
Query: 106 SDDGGGSGGGGGGG 119
S GGG GGGGGGG
Sbjct: 322 SRGGGGGGGGGGGG 363
Score = 26.6 bits (57), Expect = 1.8
Identities = 8/14 (57%), Positives = 9/14 (64%)
Frame = +3
Query: 84 WWGRRRQWWRWRQW 97
WW RR+ WRW W
Sbjct: 327 WWRWRRRRWRWWIW 368
Score = 24.6 bits (52), Expect = 6.7
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = +1
Query: 100 DGGGSGSDDGGGSGGGGGGGG 120
DG ++ GGGGGGGG
Sbjct: 289 DGRNITVNEAQSRGGGGGGGG 351
Score = 24.3 bits (51), Expect(2) = 7e-07
Identities = 6/11 (54%), Positives = 7/11 (63%)
Frame = +3
Query: 30 WCWWRRWRRSV 40
W WW RW R +
Sbjct: 390 WWWWSRWWRGI 422
>TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wall structural
protein 1.0 precursor (GRP 1.0), partial (27%)
Length = 447
Score = 46.6 bits (109), Expect(2) = 2e-07
Identities = 27/58 (46%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Frame = +3
Query: 64 GGG--GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG G GGG GGS GGGAR GGG G+ G G GGG GGG
Sbjct: 84 GGGAVGGGGGGGGGSGGGGAR------------------GGGYGAGGGSGEGGGHGGG 203
Score = 43.9 bits (102), Expect = 1e-05
Identities = 24/57 (42%), Positives = 25/57 (43%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG G G Q G G G+ GGG GGGG GGG
Sbjct: 12 GAGGAHGGGYGGGEGAG---------------QGAGGGYGGGAVGGGGGGGGGSGGG 137
Score = 43.5 bits (101), Expect = 1e-05
Identities = 25/57 (43%), Positives = 26/57 (44%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG G G+GG GGGA G GGG GS GG GGG G GG
Sbjct: 42 GGGEGAGQGAGGGYGGGAVGGGG--------------GGGGGSGGGGARGGGYGAGG 170
Score = 27.7 bits (60), Expect = 0.79
Identities = 11/18 (61%), Positives = 12/18 (66%)
Frame = +3
Query: 102 GGSGSDDGGGSGGGGGGG 119
GG+G GGG GGG G G
Sbjct: 9 GGAGGAHGGGYGGGEGAG 62
Score = 22.7 bits (47), Expect(2) = 2e-07
Identities = 6/9 (66%), Positives = 6/9 (66%)
Frame = +2
Query: 29 GWCWWRRWR 37
G CWW WR
Sbjct: 8 GRCWWGTWR 34
>TC204083 similar to UP|O48567 (O48567) Glycine-rich RNA-binding protein,
partial (95%)
Length = 1019
Score = 43.1 bits (100), Expect(2) = 3e-07
Identities = 25/57 (43%), Positives = 28/57 (48%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GSGG GGG +G RR+ R +GGG G GGG G GG G
Sbjct: 332 GSGGGYNRGSGGYGGGGGGGGYGGRREGGYNR----NGGGGGGGYGGGRDRGYGGDG 490
Score = 33.5 bits (75), Expect(2) = 7e-04
Identities = 19/56 (33%), Positives = 22/56 (38%)
Frame = +2
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
+GG +GGG GG GGG +G G G GG GGG G
Sbjct: 410 EGGYNRNGGGGGGGYGGGRDRGYG--------------GDGGSRYSRGGEGGGSDG 535
Score = 30.0 bits (66), Expect = 0.16
Identities = 20/54 (37%), Positives = 22/54 (40%), Gaps = 9/54 (16%)
Frame = +2
Query: 76 SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDG---------GGSGGGGGGGG 120
S GGG +G + R GGG G G GG GGGG GGG
Sbjct: 302 SRGGGGGGGFGSGGGYNRGSGGYGGGGGGGGYGGRREGGYNRNGGGGGGGYGGG 463
Score = 25.4 bits (54), Expect(2) = 3e-07
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +1
Query: 30 WCWWRRWRR 38
W WWRRW R
Sbjct: 307 WWWWRRWFR 333
Score = 24.6 bits (52), Expect = 6.7
Identities = 6/10 (60%), Positives = 8/10 (80%)
Frame = +1
Query: 88 RRQWWRWRQW 97
R+ WW WR+W
Sbjct: 298 RQPWWWWRRW 327
Score = 23.5 bits (49), Expect(2) = 7e-04
Identities = 6/11 (54%), Positives = 7/11 (63%)
Frame = +1
Query: 30 WCWWRRWRRSV 40
W WW WRR +
Sbjct: 364 WIWWWWWRRRI 396
>TC207679 similar to UP|Q7XJI7 (Q7XJI7) Glycine-rich protein TomR2, partial
(22%)
Length = 425
Score = 47.0 bits (110), Expect = 1e-06
Identities = 26/56 (46%), Positives = 26/56 (46%)
Frame = +3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GG GGGSGG GGGA GGGSG G GSGGGG GG
Sbjct: 24 GGYGSGGGSGGGYGGGAAG---------------GGGGGSGGGGGAGSGGGGAHGG 146
Score = 47.0 bits (110), Expect(2) = 5e-07
Identities = 25/55 (45%), Positives = 26/55 (46%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGGG+ GGG GS GGGA GGG G GGG GGG GG
Sbjct: 84 GGGGSGGGGGAGSGGGGAH------------------GGGYGGGAGGGEGGGHGG 194
Score = 45.8 bits (107), Expect = 3e-06
Identities = 25/55 (45%), Positives = 26/55 (46%)
Frame = +3
Query: 65 GGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG GGS GGG G GG G GGG+GGG GGG
Sbjct: 63 GGGAAGGGGGGSGGGGGAGSGG--------------GGAHGGGYGGGAGGGEGGG 185
Score = 44.3 bits (103), Expect = 8e-06
Identities = 25/55 (45%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Frame = +3
Query: 65 GGGNDGG-GSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GG ++GG GSGG SGGG + GGG GS GGG+G GGGG
Sbjct: 9 GGAHEGGYGSGGGSGGG-------------YGGGAAGGGGGGSGGGGGAGSGGGG 134
Score = 43.5 bits (101), Expect = 1e-05
Identities = 23/56 (41%), Positives = 25/56 (44%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG+ G GGG+ G G G G GGG GGG GGG
Sbjct: 45 GSGGGYGGGAAGGGGGGSGGGGGA-------------GSGGGGAHGGGYGGGAGGG 173
Score = 40.8 bits (94), Expect = 9e-05
Identities = 24/58 (41%), Positives = 27/58 (46%), Gaps = 2/58 (3%)
Frame = +3
Query: 64 GGGGNDGGGSGG--SSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
G GG GGG GG + GGG + GGG+GS GG GGG GGG
Sbjct: 33 GSGGGSGGGYGGGAAGGGGGGS---------------GGGGGAGSGGGGAHGGGYGGG 161
Score = 32.0 bits (71), Expect = 0.042
Identities = 13/22 (59%), Positives = 14/22 (63%)
Frame = +3
Query: 99 DDGGGSGSDDGGGSGGGGGGGG 120
+ G GSG GGG GGG GGG
Sbjct: 21 EGGYGSGGGSGGGYGGGAAGGG 86
Score = 30.8 bits (68), Expect = 0.093
Identities = 20/60 (33%), Positives = 21/60 (34%)
Frame = +3
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG SGGGG GGGG GGG GG +GGG
Sbjct: 84 GGGGSGGGGGAG------------------------------SGGGGAHGGGYGGGAGGG 173
Score = 28.1 bits (61), Expect = 0.60
Identities = 11/19 (57%), Positives = 12/19 (62%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGG 119
G G + G GSGGG GGG
Sbjct: 3 GAGGAHEGGYGSGGGSGGG 59
Score = 27.3 bits (59), Expect = 1.0
Identities = 10/19 (52%), Positives = 12/19 (62%)
Frame = +3
Query: 102 GGSGSDDGGGSGGGGGGGG 120
G G+ +GG GGG GGG
Sbjct: 3 GAGGAHEGGYGSGGGSGGG 59
Score = 25.8 bits (55), Expect = 3.0
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +2
Query: 30 WCWWRRWRR 38
W WWRRW R
Sbjct: 92 WLWWRRWCR 118
Score = 24.3 bits (51), Expect = 8.7
Identities = 7/8 (87%), Positives = 7/8 (87%), Gaps = 1/8 (12%)
Frame = +2
Query: 30 WCWW-RRW 36
WCWW RRW
Sbjct: 158 WCWWWRRW 181
Score = 20.8 bits (42), Expect(2) = 5e-07
Identities = 8/14 (57%), Positives = 8/14 (57%), Gaps = 4/14 (28%)
Frame = +2
Query: 30 WCWWR----RWRRS 39
W WWR WRRS
Sbjct: 32 W*WWRIRRRLWRRS 73
>BU763328 similar to SP|P10496|GRP2_ Glycine-rich cell wall structural
protein 1.8 precursor (GRP 1.8). [Kidney bean French
bean], partial (11%)
Length = 169
Score = 48.1 bits (113), Expect = 6e-07
Identities = 25/57 (43%), Positives = 29/57 (50%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG GGGA +G + GGG G +GGG+GGG G G
Sbjct: 161 GDGGAHGGGYGGGQGGGAGGGYGAGGE---------HGGGYGGGEGGGAGGGYGASG 18
Score = 28.5 bits (62), Expect = 0.46
Identities = 12/19 (63%), Positives = 12/19 (63%)
Frame = -3
Query: 101 GGGSGSDDGGGSGGGGGGG 119
G G G GGG GGG GGG
Sbjct: 167 GYGDGGAHGGGYGGGQGGG 111
Score = 25.0 bits (53), Expect = 5.1
Identities = 7/13 (53%), Positives = 8/13 (60%)
Frame = -2
Query: 85 WGRRRQWWRWRQW 97
W R R W +WR W
Sbjct: 48 WSRWRVWCKWRTW 10
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 48.1 bits (113), Expect = 6e-07
Identities = 38/110 (34%), Positives = 41/110 (36%), Gaps = 8/110 (7%)
Frame = -1
Query: 19 VEGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGND-----GGGS 73
++GGGD GGGG E GGGG D GGG
Sbjct: 213 IQGGGDGGGGGG----------------------E*TGGGGE*TGGGGGGDK*G*TGGGG 100
Query: 74 GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGG---GGGGGG 120
GG GGG D GGG G GGG+GG GGGGGG
Sbjct: 99 GGECGGGG-----------------DGGGGGGEYVGGGAGGEYTGGGGGG 1
Score = 45.8 bits (107), Expect = 3e-06
Identities = 25/60 (41%), Positives = 26/60 (42%), Gaps = 5/60 (8%)
Frame = -1
Query: 64 GGGGNDGGGS-----GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
GGGG+ GGG GG G W G C DGGG GG GGGGGG
Sbjct: 372 GGGGDGGGGELY*IGGGGGDGDGGGW*GAE---------CGDGGGGECSSQGGGGGGGGG 220
Score = 44.3 bits (103), Expect = 8e-06
Identities = 34/102 (33%), Positives = 36/102 (34%), Gaps = 2/102 (1%)
Frame = -1
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGGD GGGW A DGGGG GG GGG
Sbjct: 324 GGGDGDGGGW*G--------------------------AECGDGGGGECSSQGGGGGGGG 223
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSD--DGGGSGGGGGGGG 120
+ G DGGG G + GGG GGGGGG
Sbjct: 222 GDSIQGG-----------GDGGGGGGE*TGGGGE*TGGGGGG 130
Score = 43.5 bits (101), Expect = 1e-05
Identities = 29/72 (40%), Positives = 30/72 (41%), Gaps = 14/72 (19%)
Frame = -1
Query: 63 DGGGGN---DGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGG---- 115
DGGGG GGG G GGG W + D GGG S GGG GGG
Sbjct: 360 DGGGGELY*IGGGGGDGDGGG-----------W*GAECGDGGGGECSSQGGGGGGGGGDS 214
Query: 116 -------GGGGG 120
GGGGG
Sbjct: 213 IQGGGDGGGGGG 178
Score = 40.4 bits (93), Expect = 1e-04
Identities = 35/108 (32%), Positives = 37/108 (33%), Gaps = 8/108 (7%)
Frame = -1
Query: 19 VEGGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSG---- 74
V GGGD GGG + GGGG DG G G
Sbjct: 375 VGGGGDGGGG------------------------------ELY*IGGGGGDGDGGGW*GA 286
Query: 75 --GSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSD--DGGGSGGGGGG 118
G GGG + G GGG G D GGG GGGGGG
Sbjct: 285 ECGDGGGGECSSQG------------GGGGGGGGDSIQGGGDGGGGGG 178
Score = 30.4 bits (67), Expect = 0.12
Identities = 28/79 (35%), Positives = 29/79 (36%), Gaps = 23/79 (29%)
Frame = -1
Query: 64 GGGGNDG------GGSGGS---SGGGARAWWGRRRQWWRWRQWCDDGGGS---------- 104
GGGG D GG G S +GGG DDGGG
Sbjct: 492 GGGGGDAYETPMTGGKGPSYTGAGGG------------------DDGGGGEL***TGVGG 367
Query: 105 GSDDGGGS----GGGGGGG 119
G D GGG GGGGG G
Sbjct: 366 GGDGGGGELY*IGGGGGDG 310
Score = 28.9 bits (63), Expect = 0.35
Identities = 14/28 (50%), Positives = 14/28 (50%), Gaps = 9/28 (32%)
Frame = -1
Query: 102 GGSGSDDGGGS---------GGGGGGGG 120
G G DDGGG GGG GGGG
Sbjct: 429 GAGGGDDGGGGEL***TGVGGGGDGGGG 346
>CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (23%)
Length = 159
Score = 47.8 bits (112), Expect = 7e-07
Identities = 25/57 (43%), Positives = 25/57 (43%)
Frame = +2
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G G G GGGGGG G
Sbjct: 44 GGGGGGGGGEGGGGGGG--------------------GGGGGXXXGEGGGGGGGGRG 154
Score = 43.1 bits (100), Expect = 2e-05
Identities = 23/57 (40%), Positives = 24/57 (41%)
Frame = +1
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG GGG G + GGGGGG G
Sbjct: 40 GGGGGXGGGGGGGGGGGG-------------------GGGGGXXEXRRRGGGGGGKG 153
Score = 40.4 bits (93), Expect = 1e-04
Identities = 22/57 (38%), Positives = 24/57 (41%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG GGG GG GGG GGG+ + G GGGG GGG
Sbjct: 45 GGGXGGGGGRGGGGGGGG-------------------GGGAXRXEKEGGGGGGEGGG 158
Score = 40.0 bits (92), Expect = 2e-04
Identities = 19/43 (44%), Positives = 23/43 (53%)
Frame = +1
Query: 78 GGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGG + G+++ GGG G GGG GGGGGGGG
Sbjct: 1 GGGRKRTTGKKQ--------AGGGGGXGGGGGGGGGGGGGGGG 105
Score = 35.4 bits (80), Expect = 0.004
Identities = 22/57 (38%), Positives = 23/57 (39%), Gaps = 6/57 (10%)
Frame = +3
Query: 70 GGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGS------GGGGGGGG 120
G G GG GGG R GGG G GGG+ GGGGGG G
Sbjct: 36 GRGGGGXGGGGGR------------------GGGGGGGGGGGAXRXEKEGGGGGGEG 152
Score = 33.5 bits (75), Expect = 0.014
Identities = 19/57 (33%), Positives = 21/57 (36%)
Frame = +2
Query: 63 DGGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
+GGGG GGG GG G +GGG GGGG GG
Sbjct: 71 EGGGGGGGGGGGGXXXG----------------------------EGGGGGGGGRGG 157
Score = 27.7 bits (60), Expect = 0.79
Identities = 18/62 (29%), Positives = 19/62 (30%)
Frame = +2
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG+ GGGG GGGG G GG GGG
Sbjct: 62 GGGEGGGGGG--------------------------------GGGGGGXXXGEGGGGGGG 145
Query: 81 AR 82
R
Sbjct: 146GR 151
Score = 27.3 bits (59), Expect = 1.0
Identities = 24/98 (24%), Positives = 25/98 (25%)
Frame = +3
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG GG GGG GGG GG GG
Sbjct: 42 GGGGXGG-------------------------------------GGGRGGGGGGGGGGGA 110
Query: 81 ARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGG 118
R +GGG GG GGG
Sbjct: 111 XRX----------------------EKEGGGGGGEGGG 158
>TC217702 similar to GB|AAL06971.1|15810091|AY056083 AT4g30260/F9N11_110
{Arabidopsis thaliana;} , partial (39%)
Length = 436
Score = 47.8 bits (112), Expect = 7e-07
Identities = 26/58 (44%), Positives = 29/58 (49%), Gaps = 2/58 (3%)
Frame = -3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRR--RQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
GGG GGG G+ G R G R+ WRWR+ D GG G DG G GG G G
Sbjct: 338 GGGAEPGGGDVGARRGRGRRDGGDGGFREGWRWREVGVDEGGRGGVDGDGDGGAWGTG 165
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 46.6 bits (109), Expect = 2e-06
Identities = 23/53 (43%), Positives = 26/53 (48%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGG 116
GGGG GGG G GGG + + + C GGG G GGG GGGG
Sbjct: 168 GGGGYGGGGGYGGGGGGGGGGCYKCGETGHIARDCSQGGGGGGRYGGGGGGGG 326
Score = 44.3 bits (103), Expect = 8e-06
Identities = 26/58 (44%), Positives = 27/58 (45%), Gaps = 1/58 (1%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSG-GGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
G GG GGG GG G GG +G GGG G G G GGGGGGGG
Sbjct: 99 GRGGYGGGGRGGRGGYGGGGGGYG--------------GGGYGGGGGYGGGGGGGGGG 230
Score = 42.7 bits (99), Expect = 2e-05
Identities = 24/57 (42%), Positives = 24/57 (42%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG G G G GGG GG G GGG GGGGGGGG
Sbjct: 114 GGGGRGGRGGYGGGGGGY--------------------GGGGYGGGGGYGGGGGGGG 224
Score = 27.3 bits (59), Expect = 1.0
Identities = 20/64 (31%), Positives = 23/64 (35%)
Frame = +3
Query: 21 GGGDSGGGGWCWWRRWRRSVVVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGGG 80
GGG GGGG C+ E + GGGG GG GG GGG
Sbjct: 201 GGGGGGGGGGCY-----------------KCGETGHIARDCSQGGGG--GGRYGGGGGGG 323
Query: 81 ARAW 84
+
Sbjct: 324 GSCY 335
Score = 23.1 bits (48), Expect(2) = 1.3
Identities = 10/20 (50%), Positives = 11/20 (55%), Gaps = 3/20 (15%)
Frame = +2
Query: 82 RAWWGRRR---QWWRWRQWC 98
R WWG RR Q W R +C
Sbjct: 302 RRWWGWRRKLLQLWGVRAFC 361
Score = 22.3 bits (46), Expect(2) = 1.3
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +2
Query: 30 WCWWRRWRR 38
W WWR WRR
Sbjct: 194 WLWWR-WRR 217
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 45.1 bits (105), Expect = 5e-06
Identities = 27/61 (44%), Positives = 27/61 (44%), Gaps = 3/61 (4%)
Frame = +3
Query: 63 DGG---GGNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGG 119
DGG GG GGG GG GGG GGG G GGG GGGG GG
Sbjct: 297 DGGRSYGGGRGGGGGGYGGGGGY------------------GGGGGYGGGGGYGGGGRGG 422
Query: 120 G 120
G
Sbjct: 423 G 425
Score = 44.7 bits (104), Expect(2) = 2e-06
Identities = 26/59 (44%), Positives = 27/59 (45%), Gaps = 2/59 (3%)
Frame = +3
Query: 64 GGGGNDGGGS--GGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGGSGGGGGGGG 120
GGGG GGG GG GGG A + R GGG GGG G GGGGG
Sbjct: 369 GGGGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGGGGG 545
Score = 40.8 bits (94), Expect(2) = 1e-05
Identities = 28/72 (38%), Positives = 30/72 (40%), Gaps = 17/72 (23%)
Frame = +3
Query: 64 GGGGNDGGGSGGS-----------------SGGGARAWWGRRRQWWRWRQWCDDGGGSGS 106
GGGG GGG GG SGGG +G R+ GGG G
Sbjct: 387 GGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRY-----GGGGGGR 551
Query: 107 DDGGGSGGGGGG 118
GGG GGGGGG
Sbjct: 552 YVGGGGGGGGGG 587
Score = 37.0 bits (84), Expect = 0.001
Identities = 26/68 (38%), Positives = 27/68 (39%), Gaps = 2/68 (2%)
Frame = +3
Query: 54 AVVVEAVVVDGGG--GNDGGGSGGSSGGGARAWWGRRRQWWRWRQWCDDGGGSGSDDGGG 111
A V+ DG G GG GG S GG R G G G G GGG
Sbjct: 234 AKAVDVTGPDGASVQGTRRGGDGGRSYGGGRGGGG-------------GGYGGGGGYGGG 374
Query: 112 SGGGGGGG 119
G GGGGG
Sbjct: 375 GGYGGGGG 398
Score = 31.2 bits (69), Expect = 0.071
Identities = 13/20 (65%), Positives = 13/20 (65%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
GG G GGG GGGGGG G
Sbjct: 291 GGDGGRSYGGGRGGGGGGYG 350
Score = 31.2 bits (69), Expect = 0.071
Identities = 21/81 (25%), Positives = 30/81 (36%), Gaps = 13/81 (16%)
Frame = +2
Query: 30 WCWWRRWRRSV----------VVVAVTVVVIVVEAVVVEAVVVDGGGGNDGGGSGGSSGG 79
W WW RWRR + ++ + + G+SG
Sbjct: 368 WWWWLRWRRWLRWWWKRRWRRCLLQLW*I--------------------------GTSG* 469
Query: 80 GA--RAWWGR-RRQWWRWRQW 97
G R WWG+ RR+WW W+ W
Sbjct: 470 GLQWRRWWGQVRRRWWWWQIW 532
Score = 28.1 bits (61), Expect(2) = 0.018
Identities = 11/18 (61%), Positives = 12/18 (66%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGA 81
GGGG GG GG GGG+
Sbjct: 537 GGGGRYVGGGGGGGGGGS 590
Score = 25.4 bits (54), Expect = 3.9
Identities = 11/20 (55%), Positives = 11/20 (55%)
Frame = +3
Query: 101 GGGSGSDDGGGSGGGGGGGG 120
G G D G GGG GGGG
Sbjct: 279 GTRRGGDGGRSYGGGRGGGG 338
Score = 25.0 bits (53), Expect = 5.1
Identities = 10/21 (47%), Positives = 11/21 (51%)
Frame = +3
Query: 64 GGGGNDGGGSGGSSGGGARAW 84
GGGG G GG GGG +
Sbjct: 534 GGGGGRYVGGGGGGGGGGSCY 596
Score = 24.3 bits (51), Expect = 8.7
Identities = 11/17 (64%), Positives = 11/17 (64%)
Frame = +3
Query: 62 VDGGGGNDGGGSGGSSG 78
V GGGG GGGS S G
Sbjct: 555 VGGGGGGGGGGSCYSCG 605
Score = 24.3 bits (51), Expect = 8.7
Identities = 9/14 (64%), Positives = 10/14 (71%)
Frame = +3
Query: 19 VEGGGDSGGGGWCW 32
V GGG GGGG C+
Sbjct: 555 VGGGGGGGGGGSCY 596
Score = 23.9 bits (50), Expect(2) = 0.018
Identities = 6/9 (66%), Positives = 7/9 (77%)
Frame = +2
Query: 30 WCWWRRWRR 38
W WW+ WRR
Sbjct: 512 WWWWQIWRR 538
Score = 21.9 bits (45), Expect(2) = 1e-05
Identities = 12/22 (54%), Positives = 13/22 (58%)
Frame = +1
Query: 39 SVVVVAVTVVVIVVEAVVVEAV 60
+V VA VVV VEAVV V
Sbjct: 346 TVEAVATVVVVATVEAVVTVVV 411
Score = 20.8 bits (42), Expect(2) = 2e-06
Identities = 6/9 (66%), Positives = 6/9 (66%)
Frame = +2
Query: 30 WCWWRRWRR 38
W WRRW R
Sbjct: 341 WLRWRRWLR 367
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.142 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,586,447
Number of Sequences: 63676
Number of extensions: 159146
Number of successful extensions: 19008
Number of sequences better than 10.0: 1697
Number of HSP's better than 10.0 without gapping: 5833
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11971
length of query: 120
length of database: 12,639,632
effective HSP length: 96
effective length of query: 24
effective length of database: 6,526,736
effective search space: 156641664
effective search space used: 156641664
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0090b.2


