

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.6
(1129 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
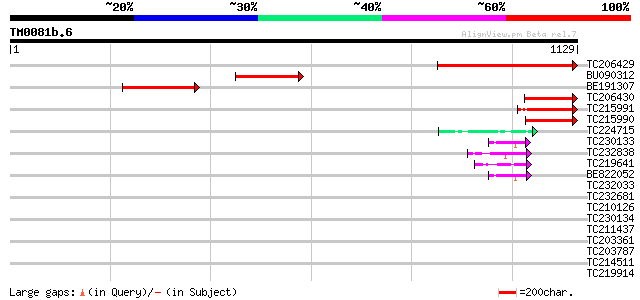
Score E
Sequences producing significant alignments: (bits) Value
TC206429 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulator... 474 e-134
BU090312 267 2e-71
BE191307 260 2e-69
TC206430 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulator... 189 8e-48
TC215991 126 6e-29
TC215990 122 9e-28
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 47 4e-05
TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 44 3e-04
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 44 3e-04
TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 43 7e-04
BE822052 similar to GP|4557063|gb|A expressed protein {Arabidops... 43 9e-04
TC232033 42 0.001
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 42 0.002
TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C2... 42 0.002
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 41 0.003
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 41 0.003
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 40 0.004
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 40 0.006
TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protei... 40 0.008
TC219914 similar to UP|Q9SAG2 (Q9SAG2) F23A5.34, partial (26%) 39 0.010
>TC206429 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulatory-like
protein, partial (23%)
Length = 1771
Score = 474 bits (1220), Expect = e-134
Identities = 238/280 (85%), Positives = 253/280 (90%), Gaps = 2/280 (0%)
Frame = +1
Query: 852 GENSLNRTNLDVSPGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDIAAPVPVANGVLVEN 911
G+NSLNRTNLDVSPG PSRPTD DDSV+KSQ VN P VEG D+A PVPVANGVL E+
Sbjct: 1 GDNSLNRTNLDVSPGRALTPSRPTDVDDSVSKSQGVNAPSVEGCDMATPVPVANGVLSES 180
Query: 912 SKVKSHEESSGPCKVEKEEGELSPNGDSEED-FVAYRDSNAQSMAKSKHNIERRKYESRD 970
SKVK+H+ES GPCK+EKEEGELSPNGDSEED VAY DSN QSMAKSKHNIERRKY+SR+
Sbjct: 181 SKVKTHDESFGPCKIEKEEGELSPNGDSEEDNIVAYGDSNVQSMAKSKHNIERRKYQSRN 360
Query: 971 REEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEED-DIEHDDVDG 1029
E+E PE GGDNDADADDEDSENVSEAGEDVSGSESAGDECF+EDHEE+ DIEHDDVDG
Sbjct: 361 GEDESCPEAGGDNDADADDEDSENVSEAGEDVSGSESAGDECFREDHEEEEDIEHDDVDG 540
Query: 1030 KAESEGEAEGMCDAQGGGDSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDSRVFYGNDD 1089
KAESEGEAEG+CDAQ GGD +SLPLSERFLSSVKPLTKHVSAVSF EEMKDSRVFYGNDD
Sbjct: 541 KAESEGEAEGICDAQAGGDGTSLPLSERFLSSVKPLTKHVSAVSFVEEMKDSRVFYGNDD 720
Query: 1090 FYALFRLHQILYERILSAKINSMSAEMKWKAKDASSPDPY 1129
FY LFRLHQ LYERILSAK +SMSAEMKWKAKDASSPDPY
Sbjct: 721 FYVLFRLHQALYERILSAKTHSMSAEMKWKAKDASSPDPY 840
>BU090312
Length = 409
Score = 267 bits (683), Expect = 2e-71
Identities = 127/136 (93%), Positives = 135/136 (98%)
Frame = +1
Query: 450 SDCKRCTPSYRLLPSDYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRKNQYEES 509
SDCKRCTPSYRLLP+DYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMR+NQYEES
Sbjct: 1 SDCKRCTPSYRLLPADYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRRNQYEES 180
Query: 510 LFRCEDDRFELDLLLESVSSASKRAEELYNNINENKISVEALSRIEDHFTVLNLRCIERL 569
LFRCEDDR+ELD+LLESVSSA+K+AEELYN+INENKISVE L+RIEDHFTVLNLRCIERL
Sbjct: 181 LFRCEDDRYELDMLLESVSSAAKKAEELYNSINENKISVETLNRIEDHFTVLNLRCIERL 360
Query: 570 YGDHGLDVIDILRKNP 585
YGDHGLDVIDILRKNP
Sbjct: 361 YGDHGLDVIDILRKNP 408
>BE191307
Length = 474
Score = 260 bits (665), Expect = 2e-69
Identities = 126/156 (80%), Positives = 140/156 (88%), Gaps = 3/156 (1%)
Frame = -2
Query: 225 RFNERNSMTPMMRQMQVDKQRYRRDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRD 284
RFNER SM PM+RQM DKQRYRRDRLPSHDRD D+S E PE+DDDKTM+N+HKEQRKR+
Sbjct: 470 RFNERGSMAPMIRQMPADKQRYRRDRLPSHDRDHDMSAERPELDDDKTMMNIHKEQRKRE 291
Query: 285 ---RRIRDQDERDPDLDNSRDLTSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDD 341
RR+RDQDER+ DLDN+RDL QRF DKKK+VKKAEGMY +AFSFCEKVKEKLSSSDD
Sbjct: 290 SRERRMRDQDEREHDLDNNRDLNLQRFPDKKKSVKKAEGMYSQAFSFCEKVKEKLSSSDD 111
Query: 342 YQTFLKCLNIFNNGIIKKNDLQNLVTDLLGKHSDLM 377
YQTFLKCL+IF+NGIIK+NDL NLVTDLLGKHSDLM
Sbjct: 110 YQTFLKCLHIFSNGIIKRNDLPNLVTDLLGKHSDLM 3
>TC206430 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulatory-like
protein, partial (14%)
Length = 1282
Score = 189 bits (479), Expect = 8e-48
Identities = 91/104 (87%), Positives = 96/104 (91%)
Frame = +1
Query: 1026 DVDGKAESEGEAEGMCDAQGGGDSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDSRVFY 1085
+ DGKAESEGEAEG+CDAQ GGD +SLPLSERFLSSVKPLTKHVSAVSF EEMKDSRVFY
Sbjct: 10 EFDGKAESEGEAEGICDAQAGGDGTSLPLSERFLSSVKPLTKHVSAVSFVEEMKDSRVFY 189
Query: 1086 GNDDFYALFRLHQILYERILSAKINSMSAEMKWKAKDASSPDPY 1129
GNDDFY FRLHQ LYER+LSAK +SMSAEMKWKAKDASSPDPY
Sbjct: 190 GNDDFYVFFRLHQALYERLLSAKTHSMSAEMKWKAKDASSPDPY 321
>TC215991
Length = 1356
Score = 126 bits (316), Expect = 6e-29
Identities = 74/120 (61%), Positives = 84/120 (69%), Gaps = 2/120 (1%)
Frame = +1
Query: 1012 CFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQG-GGDSSSLPLSERFLSSVKPLTKHVS 1070
C +E+HE D EHD+ KAESEGEAEG+ DA GD SLP SERFL +VKPL KHV
Sbjct: 1 CSREEHE--DGEHDN---KAESEGEAEGIADAHDVEGDGMSLPYSERFLLTVKPLAKHVP 165
Query: 1071 AVSFAEEMKDSRVFYGNDDFYALFRLHQILYERILSAKINSMSAEMKWKA-KDASSPDPY 1129
+ E+ ++SRVFYGND Y L RLHQ LYERI SAKINS SA+ KWKA D SS D Y
Sbjct: 166 PM-LHEKDRNSRVFYGNDSIYVLLRLHQTLYERIQSAKINSSSADRKWKASSDTSSTDQY 342
>TC215990
Length = 884
Score = 122 bits (306), Expect = 9e-28
Identities = 67/104 (64%), Positives = 74/104 (70%), Gaps = 2/104 (1%)
Frame = +2
Query: 1028 DGKAESEGEAEGMCDAQG-GGDSSSLPLSERFLSSVKPLTKHVSAVSFAEEMKDSRVFYG 1086
D KAESEGEAEGM DA GD +SLP SERFL +VKPL KHV V ++ + RVFYG
Sbjct: 8 DNKAESEGEAEGMTDANDVEGDGASLPYSERFLVTVKPLAKHVPPV-LHDKQRTVRVFYG 184
Query: 1087 NDDFYALFRLHQILYERILSAKINSMSAEMKWKA-KDASSPDPY 1129
ND FY LFRLHQ LYERI SAK+NS SAE KW+A D S D Y
Sbjct: 185 NDSFYVLFRLHQTLYERIQSAKVNSSSAEKKWRASNDTGSSDQY 316
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 47.4 bits (111), Expect = 4e-05
Identities = 54/203 (26%), Positives = 75/203 (36%), Gaps = 6/203 (2%)
Frame = -2
Query: 854 NSLNRTNLDVSPGCVSAPSRPTDADDSVAKSQTVNLPLVEGGDIAAPVPVANGVLVENSK 913
N + + D PG V P A DS K + V G P
Sbjct: 1197 NKASDSTTDERPGPVDPVVGPGPAHDSRPKCHSG----VHGRSREGPT---------QQN 1057
Query: 914 VKSHEESSGPCKVEKEEGELSPNG---DSEEDFVAYRDSNAQSMAKSKHNIERRKYESRD 970
V + E++G + L NG D + D Q +A S + E + S
Sbjct: 1056 VSADNETNGDGSNNSQITLLGVNGSCVDRVHQTEGHHDLQHQGVANSNSSGESECWNSGG 877
Query: 971 REEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEED---DIEHDDV 1027
+E G E GGD+ A+ E G+DV + GD E E D D+ DV
Sbjct: 876 GDE--GQEEGGDDGAE----------ELGDDVDDAAEEGDVAADEGAEGDGGVDVAAGDV 733
Query: 1028 DGKAESEGEAEGMCDAQGGGDSS 1050
A+ +G+ EG C GGGD +
Sbjct: 732 G--ADGDGDEEGECMGDGGGDEA 670
>TC230133 weakly similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 850
Score = 44.3 bits (103), Expect = 3e-04
Identities = 27/92 (29%), Positives = 44/92 (47%), Gaps = 9/92 (9%)
Frame = +3
Query: 954 MAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVS--EAGEDVSGSE----- 1006
+ K KH E K ++ D E++ + D + D DDE+ E+ S + GE+ +
Sbjct: 234 LCKGKHTSEENK-DASDTEDDDDDDNVNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEAN 410
Query: 1007 --SAGDECFQEDHEEDDIEHDDVDGKAESEGE 1036
D+ +D E+DD + DD D E EG+
Sbjct: 411 GGGGSDDDDGDDDEDDDNDEDDGDEDDEDEGD 506
Score = 40.8 bits (94), Expect = 0.003
Identities = 21/74 (28%), Positives = 36/74 (48%)
Frame = +3
Query: 960 NIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEE 1019
N+ + ++ D E+E G +AD+DD+ N +D G + D+ ++D +E
Sbjct: 306 NVNDGEDDNDDEEDEDFSGDDGGEEADSDDDPEANGGGGSDDDDGDDDEDDDNDEDDGDE 485
Query: 1020 DDIEHDDVDGKAES 1033
DD + D D ES
Sbjct: 486 DDEDEGDDDEDEES 527
Score = 31.2 bits (69), Expect = 2.7
Identities = 22/76 (28%), Positives = 36/76 (46%), Gaps = 2/76 (2%)
Frame = +3
Query: 976 GPETGGDND--ADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAES 1033
G T +N +D +D+D ++ GED + +D E++D DD +A+S
Sbjct: 243 GKHTSEENKDASDTEDDDDDDNVNDGEDDN-----------DDEEDEDFSGDDGGEEADS 389
Query: 1034 EGEAEGMCDAQGGGDS 1049
+ + E A GGG S
Sbjct: 390 DDDPE----ANGGGGS 425
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 44.3 bits (103), Expect = 3e-04
Identities = 36/136 (26%), Positives = 60/136 (43%), Gaps = 7/136 (5%)
Frame = +2
Query: 911 NSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRD 970
+SK+K+ +E S +VE+ E ++ D EE +S + + D
Sbjct: 212 DSKMKATKELSE--QVEENEVKVLVEEDDEE-------------YESVDEVVDDAEDDED 346
Query: 971 REEECGPETGGDNDAD-------ADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIE 1023
EE+ + G DND + DD+D E+ E G+ G E D+ +D +DD +
Sbjct: 347 EEEDDDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGDVQRGGEPDDDDNDSDDDSDDDED 526
Query: 1024 HDDVDGKAESEGEAEG 1039
D+ D + + E E G
Sbjct: 527 EDEEDEEEQGEEEDLG 574
Score = 34.3 bits (77), Expect = 0.32
Identities = 26/128 (20%), Positives = 51/128 (39%), Gaps = 4/128 (3%)
Frame = +2
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKY----ESRDREEECGPETG 980
K +E + E + + + DS ++ + +E + E D E E E
Sbjct: 143 KRREEAAQRISKSRGERNVLWFLDSKMKATKELSEQVEENEVKVLVEEDDEEYESVDEVV 322
Query: 981 GDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGM 1040
D + D D+E+ ++ E ++ + A D +D E+D+ E D G + + +
Sbjct: 323 DDAEDDEDEEEDDDDDEGDDNDDEEDDAPDGGDDDDDEDDEEEGDVQRGGEPDDDDNDSD 502
Query: 1041 CDAQGGGD 1048
D+ D
Sbjct: 503 DDSDDDED 526
>TC219641 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 942
Score = 43.1 bits (100), Expect = 7e-04
Identities = 34/114 (29%), Positives = 54/114 (46%), Gaps = 1/114 (0%)
Frame = +1
Query: 926 VEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPET-GGDND 984
+ K+ + +GD EED D +A D+++E G E G+ +
Sbjct: 364 LNKDASDSDEDGDEEED-----DDDAD-----------------DQDDEEGDEDFSGEGE 477
Query: 985 ADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAE 1038
+AD ED + AG GS+S GD+ + ++DD ++ D DG+ E EGE E
Sbjct: 478 EEADPEDDPEANGAG----GSDSDGDDD-DDGGDDDDDDNGDDDGEDEEEGEDE 624
>BE822052 similar to GP|4557063|gb|A expressed protein {Arabidopsis thaliana},
partial (28%)
Length = 668
Score = 42.7 bits (99), Expect = 9e-04
Identities = 28/92 (30%), Positives = 42/92 (45%), Gaps = 6/92 (6%)
Frame = -1
Query: 954 MAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSE------S 1007
+ K KH K ++ D +E+ E D + D+E E+ S GE+ + E
Sbjct: 566 LCKGKHTYRLNK-DASDSDEDEDEEDDHDTNDQNDEEGDEDFSGEGEEEADPEDDPEANG 390
Query: 1008 AGDECFQEDHEEDDIEHDDVDGKAESEGEAEG 1039
AG +D +EDD + +D D E E E EG
Sbjct: 389 AGGSNDDDDSDEDDDDDNDGDDDGEDEDEKEG 294
Score = 33.1 bits (74), Expect = 0.71
Identities = 19/67 (28%), Positives = 30/67 (44%)
Frame = -1
Query: 982 DNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMC 1041
D+D D D+ED + ++ ++ G E E +E EDD E + G + + E
Sbjct: 521 DSDEDEDEEDDHDTNDQNDE-EGDEDFSGEGEEEADPEDDPEANGAGGSNDDDDSDEDDD 345
Query: 1042 DAQGGGD 1048
D G D
Sbjct: 344 DDNDGDD 324
Score = 32.3 bits (72), Expect = 1.2
Identities = 20/95 (21%), Positives = 41/95 (43%)
Frame = -1
Query: 925 KVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDND 984
++ K+ + + D E+D D+N Q+ + + E D E++ G ++
Sbjct: 542 RLNKDASDSDEDEDEEDD----HDTNDQNDEEGDEDFSGEGEEEADPEDDPEANGAGGSN 375
Query: 985 ADADDEDSENVSEAGEDVSGSESAGDECFQEDHEE 1019
D D ++ ++ G+D E + +ED EE
Sbjct: 374 DDDDSDEDDDDDNDGDDDGEDEDEKEGEDEEDEEE 270
>TC232033
Length = 865
Score = 42.4 bits (98), Expect = 0.001
Identities = 61/265 (23%), Positives = 99/265 (37%), Gaps = 10/265 (3%)
Frame = +2
Query: 248 RDRLPSHDRDRDLSVEHPEMDDDKTMINLHKEQRKRDRRIRDQDER------DPDLDNSR 301
RDR D+DRD EH D L + + D R+ ++ E D D D++
Sbjct: 23 RDRSRDRDQDRDRRFEHMHDYDQAREAMLENDWIREDDRVENEQEHSRGHGGDVDRDHNI 202
Query: 302 DLTSQRFRDKKKTVKKAEGMYGEAFSFCEKVKEKLSSSDDYQTFLKCLNIFNNGIIKKND 361
DL R D+ + SF E K++ ++D LN+ N I+
Sbjct: 203 DLNVDRKMDRTSDPDR---------SFGEDRKDQSRRNND-------LNVTNQHIM---- 322
Query: 362 LQNLVTDLLGKHSDLMDEFKDFLERCENIEGFLAGVMSKKSLSTDAHLSRSSKLEDKDKD 421
+L +D G H++ + + ++ LE +K L +S KLED
Sbjct: 323 --DLSSDSAGTHNEQVSQMEEGLE--------------EKRLHVKELQKQSKKLED---- 442
Query: 422 QKREMDGAKEKDRYKEKYMGKSIQELDLSDCKRCTPSYRLLPSDYPIPTASQRSELGAQV 481
+ AK+ Y++ + K K CT + + + SE+
Sbjct: 443 ---AVINAKKNSSYRQMQLMK--LHKGFLQVKDCTEKLKTSEKELQALIDTAMSEIDCDG 607
Query: 482 LNDHWVSVTSGSED----YSFKHMR 502
L D +T+GS D Y F+ R
Sbjct: 608 LKDG--QLTNGSLDGRL*YIFRRFR 676
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 42.0 bits (97), Expect = 0.002
Identities = 34/112 (30%), Positives = 49/112 (43%), Gaps = 25/112 (22%)
Frame = +1
Query: 967 ESRDREEECGPETGG-----DNDADADDEDSENVSEAGED------------VSGSESAG 1009
+ D EEE + GG DND D +DED E+ E GE+ V+ E
Sbjct: 283 DDEDEEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQGEEADLGTEYLIRPLVTAEEEEA 462
Query: 1010 DECF--QEDHEEDDIEHDDVDG-KAES-----EGEAEGMCDAQGGGDSSSLP 1053
F +E+ EE++ + DD DG K+E+ + + D GG D P
Sbjct: 463 SSDFEPEENGEEEEEDVDDEDGEKSEAPPKRKRSDKDDSDDDDGGEDDDERP 618
Score = 38.9 bits (89), Expect = 0.013
Identities = 20/61 (32%), Positives = 31/61 (50%)
Frame = +1
Query: 967 ESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDD 1026
+ D +E+ P+ GGD+D D D+E+ +V GE DE E+ EE+ E D
Sbjct: 232 DEEDDDEDDAPD-GGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQGEEAD 408
Query: 1027 V 1027
+
Sbjct: 409 L 411
Score = 37.4 bits (85), Expect = 0.038
Identities = 20/73 (27%), Positives = 35/73 (47%)
Frame = +1
Query: 982 DNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAESEGEAEGMC 1041
D++ D +D+D ++ + G+D + + Q E DD ++DD D + E E E
Sbjct: 220 DDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQGE 399
Query: 1042 DAQGGGDSSSLPL 1054
+A G + PL
Sbjct: 400 EADLGTEYLIRPL 438
Score = 32.0 bits (71), Expect = 1.6
Identities = 18/72 (25%), Positives = 35/72 (48%), Gaps = 2/72 (2%)
Frame = +1
Query: 963 RRKYESRDREEECGPETGGDNDAD--ADDEDSENVSEAGEDVSGSESAGDECFQEDHEED 1020
RR+ D ++ E ++DA DD+D E+ E + G E D+ +D +ED
Sbjct: 193 RRRLSFDDDDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDED 372
Query: 1021 DIEHDDVDGKAE 1032
+ + ++ +A+
Sbjct: 373 EEDEEEQGEEAD 408
>TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23), partial
(7%)
Length = 735
Score = 41.6 bits (96), Expect = 0.002
Identities = 25/70 (35%), Positives = 32/70 (45%), Gaps = 13/70 (18%)
Frame = +3
Query: 982 DNDADADDEDSENVSEAGEDVSGSESAGDE-------------CFQEDHEEDDIEHDDVD 1028
D D D DDED + + GED S GDE +D +++D E DD D
Sbjct: 348 DKDGDEDDEDGHDQDDDGEDEEFSGEEGDEEGDPEDDPEANGGGSDDDDDDNDNEEDDDD 527
Query: 1029 GKAESEGEAE 1038
G+ E E E E
Sbjct: 528 GEDEEEEEDE 557
Score = 35.4 bits (80), Expect = 0.14
Identities = 23/67 (34%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Frame = +3
Query: 962 ERRKYESRDREEECGPE-----TGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQED 1016
E ++ + +EE PE GG +D D DD D+E + GED E +ED
Sbjct: 402 EDEEFSGEEGDEEGDPEDDPEANGGGSDDDDDDNDNEEDDDDGED---------EEEEED 554
Query: 1017 HEEDDIE 1023
EED+ E
Sbjct: 555 EEEDEEE 575
Score = 29.6 bits (65), Expect = 7.9
Identities = 19/57 (33%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Frame = +3
Query: 993 ENVSEAGEDVSGSESAGDECFQEDHEEDDI-EHDDVDG-KAESEGEAEGMCDAQGGG 1047
EN + ED + GDE ++ H++DD E ++ G + + EG+ E +A GGG
Sbjct: 318 ENKNSETED---DDKDGDEDDEDGHDQDDDGEDEEFSGEEGDEEGDPEDDPEANGGG 479
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 41.2 bits (95), Expect = 0.003
Identities = 28/89 (31%), Positives = 39/89 (43%), Gaps = 6/89 (6%)
Frame = +2
Query: 956 KSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVS------EAGEDVSGSESAG 1009
K KH E K S +++ + D D DDED E+ S EA D + G
Sbjct: 236 KGKHTSEENKDASDTEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEADSDDDPEANGG 415
Query: 1010 DECFQEDHEEDDIEHDDVDGKAESEGEAE 1038
E +D +EDD DD D E +G+ +
Sbjct: 416 GE--SDDDDEDD--DDDDDDNDEDDGDED 490
Score = 33.5 bits (75), Expect = 0.55
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 2/76 (2%)
Frame = +2
Query: 976 GPETGGDND--ADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDDVDGKAES 1033
G T +N +D +D+D ++ GED GD+ +D +++D DD +A+S
Sbjct: 239 GKHTSEENKDASDTEDDDDDDDVNDGED-------GDD---DDEDDEDFSGDDGGEEADS 388
Query: 1034 EGEAEGMCDAQGGGDS 1049
+ + E A GGG+S
Sbjct: 389 DDDPE----ANGGGES 424
Score = 30.4 bits (67), Expect = 4.6
Identities = 26/117 (22%), Positives = 47/117 (39%), Gaps = 6/117 (5%)
Frame = +2
Query: 891 LVEGGDIAAPVPVANGVLVENSKVK-SHEESSGPCKVEKEEGELSPNG-----DSEEDFV 944
L + ++ +P+ L + K K + EE+ E ++ + N D +ED
Sbjct: 170 LTDANNLVSPMVDGRFPLDQPCKGKHTSEENKDASDTEDDDDDDDVNDGEDGDDDDEDDE 349
Query: 945 AYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAGED 1001
+ + A S + E D ++E + DND D DED ++ + ED
Sbjct: 350 DFSGDDGGEEADSDDDPEANGGGESDDDDEDDDDDDDDNDEDDGDEDDDDEEDEDED 520
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 41.2 bits (95), Expect = 0.003
Identities = 28/85 (32%), Positives = 38/85 (43%), Gaps = 6/85 (7%)
Frame = +1
Query: 960 NIERRKYESRDREEECGPETGGDNDADADDEDSENVSEAG------EDVSGSESAGDECF 1013
N E E D ++E GP D D D +D E E G ED + E + D
Sbjct: 358 NSETEDEEDGDEDDEDGP------DEDEDGDDEEFSGEEGDEGGDPEDEANGEGSDDGDE 519
Query: 1014 QEDHEEDDIEHDDVDGKAESEGEAE 1038
+D +++ E DD DG+ E E E E
Sbjct: 520 DDDGDDNGDEEDDDDGEDEEEDEEE 594
Score = 30.0 bits (66), Expect = 6.0
Identities = 22/61 (36%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Frame = +1
Query: 967 ESRDREEECGPE--TGGDNDADADDEDSENVSEA--GEDVSGSESAGDECFQEDHEEDDI 1022
E D EE G E GGD + +A+ E S++ E G+D E D +E+ EE+D
Sbjct: 421 EDGDDEEFSGEEGDEGGDPEDEANGEGSDDGDEDDDGDDNGDEEDDDDGEDEEEDEEEDE 600
Query: 1023 E 1023
E
Sbjct: 601 E 603
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 40.4 bits (93), Expect = 0.004
Identities = 31/109 (28%), Positives = 50/109 (45%), Gaps = 14/109 (12%)
Frame = -1
Query: 929 EEGELSPNGD----------SEEDFVAYRDSNAQSMAKSKHNIER----RKYESRDREEE 974
+EG+ +GD S ED Y +N+ + + SK E + + E+E
Sbjct: 577 DEGDDDDDGDGGFGEGEEELSSEDGGGY-GNNSNNKSNSKKAPEGGAGGAEENGEEDEDE 401
Query: 975 CGPETGGDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIE 1023
G + D D D DD+D E+ ED G+E +E +ED EE+ ++
Sbjct: 400 DGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEEEEALQ 254
Score = 38.9 bits (89), Expect = 0.013
Identities = 26/117 (22%), Positives = 45/117 (38%), Gaps = 32/117 (27%)
Frame = -1
Query: 964 RKYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAG-------------- 1009
RK +S+D++ + E G D++ D DD+ E E++S + G
Sbjct: 634 RKTQSQDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKKA 455
Query: 1010 ------------------DECFQEDHEEDDIEHDDVDGKAESEGEAEGMCDAQGGGD 1048
D Q+D E+DD + DD + + E + +G D + D
Sbjct: 454 PEGGAGGAEENGEEDEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEENEED 284
Score = 37.0 bits (84), Expect = 0.049
Identities = 23/91 (25%), Positives = 42/91 (45%), Gaps = 1/91 (1%)
Frame = -1
Query: 937 GDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADA-DDEDSENV 995
G+ EE+ + + + +K N ++ EE G E ++ D DDED ++
Sbjct: 538 GEGEEELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDEDDDDD 359
Query: 996 SEAGEDVSGSESAGDECFQEDHEEDDIEHDD 1026
+ ED G E D E++EED+ + ++
Sbjct: 358 DDDEEDEGGEEEDEDGAEDEENEEDEEDEEE 266
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 40.0 bits (92), Expect = 0.006
Identities = 22/89 (24%), Positives = 42/89 (46%)
Frame = -3
Query: 937 GDSEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPETGGDNDADADDEDSENVS 996
G+ EE+ + + + +K N ++ EE G E ++ D DD+D ++
Sbjct: 571 GEGEEELSSEDGGGYGNNSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDD 392
Query: 997 EAGEDVSGSESAGDECFQEDHEEDDIEHD 1025
+ ED +G E D E++EED+ + +
Sbjct: 391 DDEEDEAGEEEDEDGAEDEENEEDEEDEE 305
Score = 39.7 bits (91), Expect = 0.008
Identities = 32/104 (30%), Positives = 50/104 (47%), Gaps = 11/104 (10%)
Frame = -3
Query: 929 EEGELSPNGD----------SEEDFVAYRDSNAQSMAKSKHNIERRKYESRDREEECGPE 978
+EG+ +GD S ED Y +N+ + + SK E + + EE E
Sbjct: 610 DEGDDDDDGDGGFGEGEEELSSEDGGGY-GNNSNNKSNSKKAPEGGAGGAEENGEEDEDE 434
Query: 979 TGGD-NDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDD 1021
G D +D D DD+D + EAGE+ + A DE +ED E+++
Sbjct: 433 DGDDQDDDDEDDDDDDEEDEAGEE-EDEDGAEDEENEEDEEDEE 305
Score = 37.4 bits (85), Expect = 0.038
Identities = 22/96 (22%), Positives = 39/96 (39%), Gaps = 22/96 (22%)
Frame = -3
Query: 965 KYESRDREEECGPETGGDNDADADDEDSENVSEAGEDVSGSESAG--------------- 1009
K +S D++ + E G D++ D DD+ E E++S + G
Sbjct: 664 KIQSEDKQRDAEGEDGNDDEGDDDDDGDGGFGEGEEELSSEDGGGYGNNSNNKSNSKKAP 485
Query: 1010 -------DECFQEDHEEDDIEHDDVDGKAESEGEAE 1038
+E +ED +ED + DD D + + E +
Sbjct: 484 EGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEED 377
>TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protein [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(88%)
Length = 1085
Score = 39.7 bits (91), Expect = 0.008
Identities = 19/29 (65%), Positives = 21/29 (71%), Gaps = 1/29 (3%)
Frame = +2
Query: 20 FASSR-GDSYGQSQVPGGGGGGGGAGGGG 47
+A SR G G+SQ GGGGGGGG GGGG
Sbjct: 668 YAGSRAGSGSGRSQGRGGGGGGGGGGGGG 754
Score = 33.9 bits (76), Expect = 0.42
Identities = 15/26 (57%), Positives = 16/26 (60%)
Frame = +2
Query: 22 SSRGDSYGQSQVPGGGGGGGGAGGGG 47
S G S G+ GGGGGGGG GG G
Sbjct: 689 SGSGRSQGRGGGGGGGGGGGGGGGSG 766
Score = 29.6 bits (65), Expect = 7.9
Identities = 18/43 (41%), Positives = 21/43 (47%), Gaps = 5/43 (11%)
Frame = +2
Query: 10 SASASQFKRPFASSRGDSYGQSQVPGGGG-----GGGGAGGGG 47
S+S+S A S SY S+ G G GGGG GGGG
Sbjct: 614 SSSSSGSGGSGAGSEAGSYAGSRAGSGSGRSQGRGGGGGGGGG 742
>TC219914 similar to UP|Q9SAG2 (Q9SAG2) F23A5.34, partial (26%)
Length = 893
Score = 39.3 bits (90), Expect = 0.010
Identities = 36/133 (27%), Positives = 62/133 (46%), Gaps = 3/133 (2%)
Frame = +1
Query: 910 ENSKVKSHEESSGPCKVEKEEGELSPNGDSEEDFVAYRDSNAQSMAKSKHNIERRKYESR 969
+ K+K EE+ K +++E EL+P + EE + +A+ K +E+ + +
Sbjct: 280 QKRKLKEEEEA----KEKEKEMELNPPEEKEE-------KSDPQLAQVKVRLEKLEEAVK 426
Query: 970 DREEECGPETG---GDNDADADDEDSENVSEAGEDVSGSESAGDECFQEDHEEDDIEHDD 1026
+ E +G G+N A DDE S A D S ++SA ++ EDH +HD
Sbjct: 427 EIVVETKKHSGSNLGENQA-TDDEKKRLNSSAPRDPSTTDSASNKAVDEDHSG---KHDI 594
Query: 1027 VDGKAESEGEAEG 1039
+ K + E +G
Sbjct: 595 LKSKPKLREETKG 633
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,366,571
Number of Sequences: 63676
Number of extensions: 530251
Number of successful extensions: 9687
Number of sequences better than 10.0: 250
Number of HSP's better than 10.0 without gapping: 4009
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6743
length of query: 1129
length of database: 12,639,632
effective HSP length: 107
effective length of query: 1022
effective length of database: 5,826,300
effective search space: 5954478600
effective search space used: 5954478600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0081b.6


