

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.3
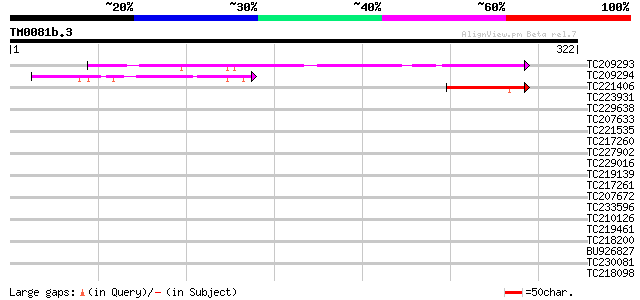
(322 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC209293 homologue to UP|RN12_HUMAN (Q9NVW2) RING finger protein... 189 1e-48
TC209294 homologue to GB|AAH21981.1|18314375|BC021981 forkhead b... 91 9e-19
TC221406 similar to UP|Q9LSS9 (Q9LSS9) General negative transcri... 61 8e-10
TC223931 homologue to UP|Q6RI84 (Q6RI84) FAR11 protein (Fragment... 40 0.002
TC229638 similar to UP|Q9EMG2 (Q9EMG2) AMV244, partial (11%) 38 0.007
TC207633 similar to UP|Q94AB7 (Q94AB7) AT5g41350/MYC6_6, partial... 37 0.016
TC221535 weakly similar to GB|AAO39941.1|28372918|BT003713 At3g0... 37 0.016
TC217260 36 0.021
TC227902 weakly similar to UP|Q8GT87 (Q8GT87) Fiber protein Fb2,... 36 0.021
TC229016 similar to UP|HES1_MOUSE (P35428) Transcription factor ... 35 0.035
TC219139 similar to UP|Q9ZT44 (Q9ZT44) RING-H2 finger protein RH... 35 0.060
TC217261 34 0.10
TC207672 weakly similar to UP|Q9ZT44 (Q9ZT44) RING-H2 finger pro... 33 0.13
TC233596 33 0.17
TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C2... 33 0.17
TC219461 homologue to UP|Q9AR19 (Q9AR19) Histone acetyltransfera... 32 0.30
TC218200 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partia... 32 0.51
BU926827 32 0.51
TC230081 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partia... 32 0.51
TC218098 weakly similar to UP|Q8K1Y5 (Q8K1Y5) Carm1-pending prot... 31 0.66
>TC209293 homologue to UP|RN12_HUMAN (Q9NVW2) RING finger protein 12 (LIM
domain interacting RING finger protein) (RING finger LIM
domain-binding protein) (R-LIM) (NY-REN-43 antigen),
partial (4%)
Length = 957
Score = 189 bits (480), Expect = 1e-48
Identities = 115/271 (42%), Positives = 142/271 (51%), Gaps = 20/271 (7%)
Frame = +3
Query: 45 VDSNGRMDNCPSS-KHTASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSP------- 96
+D +G P+ KH+ N R+GE+ +G DSDS S N P
Sbjct: 15 LDDDGHAPPSPAEVKHSVEPLNTR------RRGEEDDGLIHHDSDSESPTNCPAGVLCGT 176
Query: 97 DRSTYSHDESSGKDFSGSSSNSISTS--CSGN----------DSEEEDDDGCLDDWEAVA 144
D T SSG S SSS+S S+ CSGN D +++DDDGCLDDWEA+A
Sbjct: 177 DSGTNFTGSSSGGSSSSSSSSSSSSGGCCSGNVTEVEEEEEGDDDDDDDDGCLDDWEAMA 356
Query: 145 DALYANDKSHSVVSESPAEHEAECRYTVLEDDKNPRVDFSKADFKSEVPESKPNRRAWKP 204
DAL A+DK + +SP + N SK + VP N RAW+
Sbjct: 357 DALAADDKRENPCPDSPPVVSPS-------EGSNLGSPNSKPESDRLVPWGSGNSRAWRA 515
Query: 205 DDTLRPRCLPNLSKQRNSPLNSNWRGSRKTVPWAWQTIISQPSQCPICYEDLDVTDSSFL 264
DD RP+ LPNLSKQ + P R+ VPW PS CPIC EDLD+TD+SF+
Sbjct: 516 DDAFRPQSLPNLSKQHSMP-----NPDRRGVPWGRAPT---PSSCPICCEDLDLTDTSFM 671
Query: 265 PCSCGFHLCLFCHKKILEADARCPSCRKLYD 295
PC CGF LCLFCHK+ILE D RCP CRK Y+
Sbjct: 672 PCLCGFRLCLFCHKRILEEDGRCPGCRKPYE 764
>TC209294 homologue to GB|AAH21981.1|18314375|BC021981 forkhead box O1A {Homo
sapiens;} , partial (3%)
Length = 592
Score = 90.5 bits (223), Expect = 9e-19
Identities = 68/152 (44%), Positives = 79/152 (51%), Gaps = 24/152 (15%)
Frame = +3
Query: 13 KKKRTNGSAKLKQIKLDVRREQWLSR---VKKGC--NVDSNGRMDNCPSS----KHTASE 63
KKKRTN SAKLKQ K+D RREQWLS+ KGC VD +G P S KH+
Sbjct: 165 KKKRTNRSAKLKQYKIDARREQWLSQGAVKSKGCKDGVDDDGHAP--PPSPAVVKHSLEP 338
Query: 64 ENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTS- 122
N R+GE+ +G DSDS S N P D SG +F+GSSS S+S
Sbjct: 339 LN------TRRRGEEDDGLIHHDSDSESPSNCPAGVLCGTD--SGTNFTGSSSGGSSSSS 494
Query: 123 ------CSGNDSEEE--------DDDGCLDDW 140
CS N +EEE DDDGCLDDW
Sbjct: 495 SSSGGCCSXNVTEEEEEXEXDDHDDDGCLDDW 590
>TC221406 similar to UP|Q9LSS9 (Q9LSS9) General negative transcription
regulator-like, partial (15%)
Length = 763
Score = 60.8 bits (146), Expect = 8e-10
Identities = 23/53 (43%), Positives = 33/53 (61%), Gaps = 6/53 (11%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILE------ADARCPSCRKLYD 295
CP+C E++D+TD PC CG+ +C++C ILE + RCP+CR YD
Sbjct: 91 CPLCAEEMDLTDQQLKPCKCGYEICVWCWHHILEMAEKDDTEGRCPACRSPYD 249
>TC223931 homologue to UP|Q6RI84 (Q6RI84) FAR11 protein (Fragment), partial
(4%)
Length = 525
Score = 39.7 bits (91), Expect = 0.002
Identities = 16/49 (32%), Positives = 27/49 (54%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLYDHV 297
C ICY + V+ + + C C H C C + + ++RCP CR+ + +V
Sbjct: 220 CGICYAESGVSIAGEIDC-CSHHFCFVCIMEWAKHESRCPICRQRFSNV 363
>TC229638 similar to UP|Q9EMG2 (Q9EMG2) AMV244, partial (11%)
Length = 780
Score = 37.7 bits (86), Expect = 0.007
Identities = 29/106 (27%), Positives = 47/106 (43%), Gaps = 19/106 (17%)
Frame = +2
Query: 205 DDTLRPRCLPNLSKQRNSP-----------LNSNWRGSRKT-------VPWAWQTIISQP 246
DD R + + +Q N+ L++ GS+K+ W+W+ S+
Sbjct: 275 DDKFRAQRMSENKRQNNTKCVEGKRTSIIVLHTEEYGSKKSGSRRFSWTKWSWKA--SEQ 448
Query: 247 SQCPICYEDLDVTDSSF-LPCSCGFHLCLFCHKKILEADARCPSCR 291
+C +C E V ++ LPC+ FH C K LE ++ CP CR
Sbjct: 449 EECAVCLESFRVGETLIHLPCAHRFH--DRCLKPWLENNSYCPCCR 580
>TC207633 similar to UP|Q94AB7 (Q94AB7) AT5g41350/MYC6_6, partial (33%)
Length = 825
Score = 36.6 bits (83), Expect = 0.016
Identities = 19/72 (26%), Positives = 34/72 (46%), Gaps = 1/72 (1%)
Frame = +3
Query: 242 IISQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSC-RKLYDHVDGN 300
++ + CPIC E+ D ++ L +C H L C + +E CP C + L
Sbjct: 591 LVEEEDACPICLEEYD-AENPKLATNCDHHFHLACILEWMERSETCPVCDQDLVFDPPIE 767
Query: 301 VGLNIGAKAFCI 312
+N+G + +C+
Sbjct: 768 *YINVGVQIYCV 803
>TC221535 weakly similar to GB|AAO39941.1|28372918|BT003713 At3g07200
{Arabidopsis thaliana;} , partial (32%)
Length = 658
Score = 36.6 bits (83), Expect = 0.016
Identities = 17/45 (37%), Positives = 23/45 (50%)
Frame = +2
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKL 293
CPIC L+ S+ C C C + L A A+CP+CRK+
Sbjct: 179 CPICMSALEEETST----RCAHIFCKNCIRAALSAQAKCPTCRKV 301
>TC217260
Length = 1114
Score = 36.2 bits (82), Expect = 0.021
Identities = 28/89 (31%), Positives = 39/89 (43%), Gaps = 5/89 (5%)
Frame = +3
Query: 209 RPRCLPNLSKQRNSPLN-----SNWRGSRKTVPWAWQTIISQPSQCPICYEDLDVTDSSF 263
R R LPNL + +N S+ GS K P + ++ CPIC + S+
Sbjct: 381 RRRELPNLIINCDKYINLEGSSSSMGGSVKKTPEPPKELVFN---CPICMGPMVHEMST- 548
Query: 264 LPCSCGFHLCLFCHKKILEADARCPSCRK 292
CG C C K + A +CP+CRK
Sbjct: 549 ---RCGHIFCKDCIKAAISAQGKCPTCRK 626
>TC227902 weakly similar to UP|Q8GT87 (Q8GT87) Fiber protein Fb2, partial
(33%)
Length = 911
Score = 36.2 bits (82), Expect = 0.021
Identities = 23/87 (26%), Positives = 42/87 (47%), Gaps = 6/87 (6%)
Frame = -2
Query: 88 DSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGNDSEEEDDDGCLDDWEAVADAL 147
D +S + R +S S KD S +N++S CS + ++ GC + ADA
Sbjct: 583 DIKSPCTNLLRCAFSIWSLSDKDG*TSLTNTLSFECSSSMLTSREESGCTFLRSSTADAP 404
Query: 148 YANDKSHSV------VSESPAEHEAEC 168
+ ND S+ ++++P E++ +C
Sbjct: 403 HKNDSHGSLAALEVDIADNPVENDCQC 323
>TC229016 similar to UP|HES1_MOUSE (P35428) Transcription factor HES-1 (Hairy
and enhancer of split 1), partial (6%)
Length = 769
Score = 35.4 bits (80), Expect = 0.035
Identities = 17/44 (38%), Positives = 22/44 (49%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CPIC L S+ CG C C + + A A+CP+CRK
Sbjct: 415 CPICMSPLVEEMST----RCGHIFCKNCIRAAISAQAKCPTCRK 534
>TC219139 similar to UP|Q9ZT44 (Q9ZT44) RING-H2 finger protein RHB1a, partial
(53%)
Length = 1200
Score = 34.7 bits (78), Expect = 0.060
Identities = 17/51 (33%), Positives = 24/51 (46%)
Frame = +1
Query: 240 QTIISQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSC 290
+ + + CPIC E+ DV + S L C H L C + +E CP C
Sbjct: 550 ELVTEEEDVCPICLEEYDVENPSNL-TKCEHHFHLSCILEWMERSDSCPIC 699
>TC217261
Length = 1186
Score = 33.9 bits (76), Expect = 0.10
Identities = 26/88 (29%), Positives = 37/88 (41%), Gaps = 4/88 (4%)
Frame = +3
Query: 209 RPRCLPNLSKQRNSPLNSNWRGSRKTVPWAWQTIISQPSQ----CPICYEDLDVTDSSFL 264
R R LPNL N N G ++ +++ P + CPIC + S+
Sbjct: 402 RRRELPNLII--NCDKYINLEGGSSSMEQSFKKPPEPPKELVFNCPICMGPMVHEMST-- 569
Query: 265 PCSCGFHLCLFCHKKILEADARCPSCRK 292
CG C C K + A +CP+CRK
Sbjct: 570 --RCGHIFCKDCIKAAISAQGKCPTCRK 647
>TC207672 weakly similar to UP|Q9ZT44 (Q9ZT44) RING-H2 finger protein RHB1a,
partial (82%)
Length = 1246
Score = 33.5 bits (75), Expect = 0.13
Identities = 16/51 (31%), Positives = 23/51 (44%)
Frame = +2
Query: 242 IISQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
+ + CPIC E+ DV + L C H L C + +E CP C +
Sbjct: 551 VTEEEDGCPICLEEYDVENPKTL-TKCEHHFHLSCILEWMERSDSCPICNQ 700
>TC233596
Length = 626
Score = 33.1 bits (74), Expect = 0.17
Identities = 16/40 (40%), Positives = 19/40 (47%), Gaps = 2/40 (5%)
Frame = -2
Query: 231 SRKTVPWA--WQTIISQPSQCPICYEDLDVTDSSFLPCSC 268
S K PW W + QP +CP C + L S LPC C
Sbjct: 613 SGKGCPWLTRWMNLKFQPEKCPSCQQCLISCHYSLLPC*C 494
>TC210126 similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23), partial
(7%)
Length = 735
Score = 33.1 bits (74), Expect = 0.17
Identities = 26/92 (28%), Positives = 39/92 (42%), Gaps = 3/92 (3%)
Frame = +3
Query: 47 SNGR--MDNCPSSKHTASEENRSSCKEMTRKG-EDIEGTCIQDSDSRSTINSPDRSTYSH 103
+NGR +++ KH E S ++ + G ED E QD D S +
Sbjct: 264 ANGRFPLEDLFGRKHAYLENKNSETEDDDKDGDEDDEDGHDQDDDGEDEEFSGEEGDEEG 443
Query: 104 DESSGKDFSGSSSNSISTSCSGNDSEEEDDDG 135
D + +G S+ ND+EE+DDDG
Sbjct: 444 DPEDDPEANGGGSDDDDDD---NDNEEDDDDG 530
Score = 28.1 bits (61), Expect = 5.6
Identities = 24/84 (28%), Positives = 38/84 (44%), Gaps = 3/84 (3%)
Frame = -3
Query: 42 GCNVDSNGRMDNCPSSKHTASEENRSSCKEMTRKGEDIEGTCIQDSDS---RSTINSPDR 98
GC V S+ + SS ++S + SS ++ + S S S+ +SP+
Sbjct: 586 GCEVSSSSSSSS--SSSSSSSPSSSSSSLSLSSSSSSLPPPLASGSSSGSPSSSPSSPEN 413
Query: 99 STYSHDESSGKDFSGSSSNSISTS 122
S+ S S S SSS S+S+S
Sbjct: 412 SSSSPSSS*SCPSSSSSSPSLSSS 341
>TC219461 homologue to UP|Q9AR19 (Q9AR19) Histone acetyltransferase GCN5
(At3g54610) (Expressed protein), partial (12%)
Length = 853
Score = 32.3 bits (72), Expect = 0.30
Identities = 30/106 (28%), Positives = 41/106 (38%), Gaps = 2/106 (1%)
Frame = +1
Query: 50 RMDNCPSSKHTASEENRSSCKEMTRKGEDIEGTCIQDSDSRSTINSPD--RSTYSHDESS 107
R PS H+AS SS + ED ++P S++S D
Sbjct: 148 RSSQSPSPSHSASASATSSIHKRKLVSED---------------HAPPFPPSSFSADTRD 282
Query: 108 GKDFSGSSSNSISTSCSGNDSEEEDDDGCLDDWEAVADALYANDKS 153
G S SIS + +DS+ E +D +DD E D ND S
Sbjct: 283 GALTSNDDLESISARGADSDSDSESEDAVVDDDEDDYDNDNDNDSS 420
>TC218200 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partial (25%)
Length = 1175
Score = 31.6 bits (70), Expect = 0.51
Identities = 13/44 (29%), Positives = 21/44 (47%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CPIC +DL + ++ CG ++ C + + CP C K
Sbjct: 643 CPICCDDLFTSSATVRALPCGHYMHSACFQAYTCSHYTCPICSK 774
>BU926827
Length = 445
Score = 31.6 bits (70), Expect = 0.51
Identities = 16/43 (37%), Positives = 18/43 (41%)
Frame = +3
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCR 291
CP+C E LD L C C K AD+ CP CR
Sbjct: 24 CPVCLERLDQDTGGILTTICNHSFHCSCISK--WADSSCPVCR 146
>TC230081 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partial (26%)
Length = 1151
Score = 31.6 bits (70), Expect = 0.51
Identities = 13/44 (29%), Positives = 21/44 (47%)
Frame = +1
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRK 292
CPIC +DL + ++ CG ++ C + + CP C K
Sbjct: 775 CPICCDDLFTSSATVRALPCGHYMHSSCFQAYTCSHYTCPICSK 906
>TC218098 weakly similar to UP|Q8K1Y5 (Q8K1Y5) Carm1-pending protein, partial
(6%)
Length = 1369
Score = 31.2 bits (69), Expect = 0.66
Identities = 19/60 (31%), Positives = 29/60 (47%), Gaps = 3/60 (5%)
Frame = +2
Query: 251 ICYEDLDVTDSSFLP---CSCGFHLCLFCHKKILEADARCPSCRKLYDHVDGNVGLNIGA 307
+ Y VTD+ +P CS +LC + +A CP C KLY ++ V N+G+
Sbjct: 602 VAYHCSWVTDNPLVPVALCSLSANLCHGRTRNYWQASLDCPQCTKLY-NILNTVS*NVGS 778
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.129 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,305,615
Number of Sequences: 63676
Number of extensions: 279915
Number of successful extensions: 1663
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 1627
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1645
length of query: 322
length of database: 12,639,632
effective HSP length: 97
effective length of query: 225
effective length of database: 6,463,060
effective search space: 1454188500
effective search space used: 1454188500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0081b.3


