

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.1
(360 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
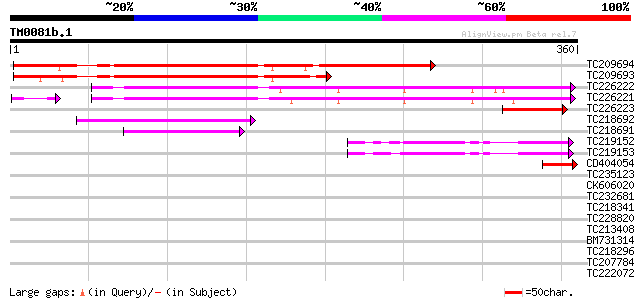
Score E
Sequences producing significant alignments: (bits) Value
TC209694 similar to UP|Q8LGA2 (Q8LGA2) Seed maturation-like prot... 329 1e-90
TC209693 similar to UP|Q8LGA2 (Q8LGA2) Seed maturation-like prot... 220 9e-58
TC226222 homologue to UP|Q9SEL1 (Q9SEL1) Seed maturation protein... 184 5e-47
TC226221 homologue to UP|Q9SEL1 (Q9SEL1) Seed maturation protein... 175 2e-44
TC226223 similar to UP|Q9SEL1 (Q9SEL1) Seed maturation protein P... 58 8e-09
TC218692 similar to UP|Q9SH45 (Q9SH45) F2K11.3, partial (47%) 57 1e-08
TC218691 similar to UP|Q9SH45 (Q9SH45) F2K11.3, partial (35%) 54 1e-07
TC219152 50 1e-06
TC219153 similar to UP|Q97HY8 (Q97HY8) Uncharacterized secreted ... 49 4e-06
CD404054 46 2e-05
TC235123 similar to GB|CAD40675.1|21741499|OSJN00124 OSJNBb0118P... 31 1.00
CK606020 27 1.1
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 28 1.6
TC218341 homologue to UP|Q6IEK9 (Q6IEK9) WRKY transcription fact... 30 1.7
TC228820 weakly similar to UP|Q8LRM1 (Q8LRM1) Nam-like protein 4... 30 2.2
TC213408 similar to UP|Q9Y1M2 (Q9Y1M2) P68 RNA helicase, partial... 30 2.2
BM731314 30 2.2
TC218296 29 2.9
TC207784 similar to GB|AAM19935.1|20453393|AY097419 AT5g58640/mz... 29 2.9
TC222072 28 4.9
>TC209694 similar to UP|Q8LGA2 (Q8LGA2) Seed maturation-like protein, partial
(40%)
Length = 810
Score = 329 bits (843), Expect = 1e-90
Identities = 191/283 (67%), Positives = 211/283 (74%), Gaps = 15/283 (5%)
Frame = +2
Query: 3 ALSWRPFILSRLTDLSPNPLHPPKPPPL-----------FLRRRRCFLTSCYADGFSSSS 51
ALS+RP I R+TD S PPKPPPL RRR FL SC
Sbjct: 20 ALSFRPVIHPRVTDRSLFQFLPPKPPPLPTSPPPSVLLPLTRRRGGFLPSCL-------- 175
Query: 52 SSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGL 111
SDD VSTR FDRGFTVIA MLRRIEPLDNS ISKGVS AARDSMKQTISTMLGL
Sbjct: 176 ----SDDAVSTRN--FDRGFTVIAAMLRRIEPLDNSAISKGVSPAARDSMKQTISTMLGL 337
Query: 112 LPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARD--SDCE 169
LPSDHF+VT+TVSKHPLHRLL SSI+TGYTLWNAEYRMSLTRNLDI+ G+RD SDCE
Sbjct: 338 LPSDHFAVTITVSKHPLHRLLFSSIVTGYTLWNAEYRMSLTRNLDIS---GSRDEGSDCE 508
Query: 170 KRSEILEVKGGGEDGGE--IEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELT 227
RSE+LEVK G + G+ IEV +DL + +CS + + FGDLPPQAL+YIQQLQSELT
Sbjct: 509 TRSEVLEVKDGAKTVGDEKIEVVNDL---ESCSCSGNLKEFGDLPPQALSYIQQLQSELT 679
Query: 228 NVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSR 270
+V EELNA+K EMMQLEYD+G NNL+EYLRSLDP MVTELSR
Sbjct: 680 SVTEELNAQKHEMMQLEYDKGKWNNLVEYLRSLDPNMVTELSR 808
>TC209693 similar to UP|Q8LGA2 (Q8LGA2) Seed maturation-like protein, partial
(25%)
Length = 646
Score = 220 bits (560), Expect = 9e-58
Identities = 133/215 (61%), Positives = 150/215 (68%), Gaps = 13/215 (6%)
Frame = +1
Query: 3 ALSWRPFILSRLTDLS--------PNPLHPPKPPPLFL---RRRRCFLTSCYADGFSSSS 51
ALS+RP I R TD S P P PP PP + L RRR F++SC
Sbjct: 46 ALSFRPLIHPRFTDRSLFQFLPPKPPPFPPPPPPSVLLPLTRRRGGFISSCL-------- 201
Query: 52 SSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGL 111
SDD VSTR FDRGFTVIA MLRRIEPLDNS ISKGVS AARDSMKQTISTMLGL
Sbjct: 202 ----SDDAVSTRN--FDRGFTVIAAMLRRIEPLDNSAISKGVSPAARDSMKQTISTMLGL 363
Query: 112 LPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARD--SDCE 169
LPSDHF+VTVTVSKHPLHRLL SSI+TGYTLWNAEYRMSL RNLD++ G+R+ SDCE
Sbjct: 364 LPSDHFAVTVTVSKHPLHRLLFSSIVTGYTLWNAEYRMSLARNLDMS---GSRNEGSDCE 534
Query: 170 KRSEILEVKGGGEDGGEIEVASDLGLKDLENCSSS 204
SE+LEVK + G+ ++ + DLE+CSSS
Sbjct: 535 TCSEVLEVKDVAKTVGDEKIDV---VNDLESCSSS 630
>TC226222 homologue to UP|Q9SEL1 (Q9SEL1) Seed maturation protein PM23
(Fragment), partial (92%)
Length = 1616
Score = 184 bits (467), Expect = 5e-47
Identities = 125/327 (38%), Positives = 184/327 (56%), Gaps = 20/327 (6%)
Frame = +1
Query: 53 SSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLL 112
+SSS D S K + V+ +++ IEPLD S I K V D+MK+TIS MLGLL
Sbjct: 208 ASSSHDFTSNSKKS------VLTELIQEIEPLDVSHIQKDVPPTTTDAMKRTISGMLGLL 369
Query: 113 PSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEK-- 170
PSD F V + PL +LL+SS++TGYTL N EYR+ L +NLD+ + D EK
Sbjct: 370 PSDQFHVVIEALWEPLSKLLISSMMTGYTLHNVEYRLCLEKNLDMF------EGDIEKPK 531
Query: 171 -RSEILEVKGGGEDG-GEIEVASDLGLKD-LENCSSSPRV--FGDLPPQALNYIQQLQSE 225
S ++++G D I+ D L +E + GD+ +AL YI LQS
Sbjct: 532 AESTKVDLQGLMHDSVNVIDFGRDKSLSSKVEKLHEDADIQELGDISAEALQYIFNLQSR 711
Query: 226 LTNVKEELNARKQEMMQLEYDRGI---RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQ 282
L+++K+EL+ K++ L+ + + +N+LL+YLRSL PE V +LS +S E++D I
Sbjct: 712 LSSMKKELHEVKRKSAALQMQQFVGEEKNDLLDYLRSLQPEQVAQLSEFTSPELKDTILF 891
Query: 283 LVQNILSRFF--VDDASGSFMEQSVEG--NIDNH------PDNGDEFSDTVGTSRDYLAK 332
+V +L+ + + E + G N+ N ++ +F V +RDYLA+
Sbjct: 892 VVHGLLATLSPKMHSKPSTISENTTVGATNVGNEDCAEVVENSAHQFQPVVSLTRDYLAR 1071
Query: 333 LLFWCMLLGHHLRGLENRLQLSCVVGL 359
LLFWCMLLGH+LRGLE RL L+ ++ L
Sbjct: 1072LLFWCMLLGHYLRGLECRLDLTDLLSL 1152
>TC226221 homologue to UP|Q9SEL1 (Q9SEL1) Seed maturation protein PM23
(Fragment), complete
Length = 1663
Score = 175 bits (443), Expect(2) = 2e-44
Identities = 119/327 (36%), Positives = 183/327 (55%), Gaps = 20/327 (6%)
Frame = +1
Query: 53 SSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLL 112
++SS D S K + V+ +++ IEPLD S I K V D+MK+TIS MLGLL
Sbjct: 223 AASSHDFASNSKKS------VLTELIQEIEPLDVSHIQKDVPPTTADAMKRTISGMLGLL 384
Query: 113 PSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEK-R 171
PSD F V + PL +LL+SS++TGYTL N EYR+ L +NLD+ + D EK +
Sbjct: 385 PSDQFHVVIEALWEPLSKLLISSMMTGYTLRNVEYRLCLEKNLDMF------EGDIEKPK 546
Query: 172 SEILEV---KGGGEDGGEIEVASDLGLKD-LENCSSSPRV--FGDLPPQALNYIQQLQSE 225
+E ++V + + IE + L +E + G++ +A YI LQS
Sbjct: 547 AESMKVDLQRLMHDSVNAIEFGKNKNLSSKVEKLHEEADIQELGEISAEAQQYIFNLQSR 726
Query: 226 LTNVKEELNARKQEMMQLEYDRGI---RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQ 282
L+++K+EL+ K++ L+ + + +N+LL+YLRSL PE V +LS +S E++D I
Sbjct: 727 LSSMKKELHEVKRKSAALQMQQFVGEEKNDLLDYLRSLQPEQVAQLSEFTSPELKDTILS 906
Query: 283 LVQNILSRFF--VDDASGSFMEQSVEGNIDNHPDNGDE--------FSDTVGTSRDYLAK 332
+V +L+ + + E + G + ++ E F + +RDYLA+
Sbjct: 907 VVHGLLATLSPKMHSKPSTMSENTTVGATNAGSEDCAEVLENSALQFQPVISLTRDYLAR 1086
Query: 333 LLFWCMLLGHHLRGLENRLQLSCVVGL 359
LLFWCMLLGH+LRGLE RL+L+ ++ L
Sbjct: 1087LLFWCMLLGHYLRGLECRLELTDLLSL 1167
Score = 21.9 bits (45), Expect(2) = 2e-44
Identities = 11/31 (35%), Positives = 14/31 (44%)
Frame = +2
Query: 2 AALSWRPFILSRLTDLSPNPLHPPKPPPLFL 32
++L W+PF PP PPLFL
Sbjct: 74 SSLVWQPF--------------PPSSPPLFL 124
>TC226223 similar to UP|Q9SEL1 (Q9SEL1) Seed maturation protein PM23
(Fragment), partial (25%)
Length = 889
Score = 57.8 bits (138), Expect = 8e-09
Identities = 25/41 (60%), Positives = 32/41 (77%)
Frame = +3
Query: 314 DNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQLS 354
++ +F V +RDYLA+LLFWCMLLGH+LRGLE RL L+
Sbjct: 120 NSAHQFQPVVSLTRDYLARLLFWCMLLGHYLRGLECRLDLT 242
Score = 29.6 bits (65), Expect = 2.2
Identities = 13/18 (72%), Positives = 15/18 (83%)
Frame = +1
Query: 337 CMLLGHHLRGLENRLQLS 354
CMLLG +LRGLE RL L+
Sbjct: 436 CMLLGRYLRGLECRLDLT 489
>TC218692 similar to UP|Q9SH45 (Q9SH45) F2K11.3, partial (47%)
Length = 644
Score = 57.4 bits (137), Expect = 1e-08
Identities = 30/114 (26%), Positives = 57/114 (49%)
Frame = +1
Query: 43 YADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMK 102
Y + +S+ +S+ D + ++ ++ ++P + K D+M+
Sbjct: 289 YDSSNNDNSNPASAGDSKPPNGTLSKSRREILLEYVKNVQPEFMELFVKRAPQQVVDAMR 468
Query: 103 QTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLD 156
QT++ M+G LP F+VT+T L +L+ S ++TGY NA+YR+ L L+
Sbjct: 469 QTVTNMIGTLPPQFFAVTITTVAENLAQLMYSIMMTGYMFKNAQYRLELQEGLE 630
>TC218691 similar to UP|Q9SH45 (Q9SH45) F2K11.3, partial (35%)
Length = 441
Score = 53.5 bits (127), Expect = 1e-07
Identities = 24/77 (31%), Positives = 44/77 (56%)
Frame = +1
Query: 73 VIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLL 132
++ ++ ++P + K D+M+QT++ M+G LP F+VT+T L +L+
Sbjct: 208 ILLEYVKNVQPEFMELFVKRAPQQVVDAMRQTVTNMIGTLPPQFFAVTITTVAENLAQLM 387
Query: 133 VSSIITGYTLWNAEYRM 149
S ++TGY NA+YR+
Sbjct: 388 YSIMMTGYMFKNAQYRL 438
>TC219152
Length = 743
Score = 50.4 bits (119), Expect = 1e-06
Identities = 44/144 (30%), Positives = 73/144 (50%)
Frame = +1
Query: 215 ALNYIQQLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSV 274
A YI+ L++E+ EELN + + + G +N LLEYL+SL+P + EL+ +
Sbjct: 91 AKKYIELLEAEI----EELNRQ----VGRQSSNG-QNELLEYLKSLEPRNLKELTSSAGE 243
Query: 275 EVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLL 334
+V ++ ++ +L+ V D S M+ SV TS LAKLL
Sbjct: 244 DVVFAMNTFIKRLLA---VSDPSQ--MKTSV-----------------TETSAAELAKLL 357
Query: 335 FWCMLLGHHLRGLENRLQLSCVVG 358
+W M++G+ +R +E R + V+G
Sbjct: 358 YWLMVVGYSIRNIEVRYDMERVLG 429
>TC219153 similar to UP|Q97HY8 (Q97HY8) Uncharacterized secreted protein,
homolog YXKC Bacillus subtilis, partial (6%)
Length = 684
Score = 48.9 bits (115), Expect = 4e-06
Identities = 42/144 (29%), Positives = 69/144 (47%)
Frame = +2
Query: 215 ALNYIQQLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSV 274
A YI+ L++E+ EEL+ + +N LLEYL+SL+P + EL+ +
Sbjct: 101 AKKYIELLEAEI----EELSCQVGRQTS-----NAQNELLEYLKSLEPRNLKELTSTAGE 253
Query: 275 EVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLL 334
+V ++ ++ +L+ V D S M+ SV TS LAKLL
Sbjct: 254 DVVFAMNAFIKRLLA---VSDPSQ--MKTSV-----------------TETSAAELAKLL 367
Query: 335 FWCMLLGHHLRGLENRLQLSCVVG 358
+W M++G+ +R +E R + V G
Sbjct: 368 YWLMVVGYSIRNIEVRYDMERVFG 439
>CD404054
Length = 187
Score = 46.2 bits (108), Expect = 2e-05
Identities = 21/22 (95%), Positives = 21/22 (95%)
Frame = -1
Query: 339 LLGHHLRGLENRLQLSCVVGLL 360
LLGHHLRGLENRL LSCVVGLL
Sbjct: 178 LLGHHLRGLENRLHLSCVVGLL 113
>TC235123 similar to GB|CAD40675.1|21741499|OSJN00124 OSJNBb0118P14.13
{Oryza sativa (japonica cultivar-group);} , partial
(15%)
Length = 417
Score = 30.8 bits (68), Expect = 1.00
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Frame = +2
Query: 4 LSWRPFILSRLTDLSPNPLHPP---KPPPLFLRRRRCFLTSCYADGFSSSSSSSSSDDVV 60
LS P + +R S PL P PP L + R ++ YA+ ++ SSS++ DVV
Sbjct: 29 LSSLPAVSARPIPTSAFPLRKPLFQTPPTLHFKNHRFAVSCSYAEAGVNADSSSNTIDVV 208
Query: 61 STRKS 65
+ +S
Sbjct: 209ADVRS 223
>CK606020
Length = 340
Score = 26.6 bits (57), Expect(2) = 1.1
Identities = 15/39 (38%), Positives = 23/39 (58%)
Frame = -3
Query: 40 TSCYADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANML 78
+S + SSSSSSSSSD +S+ +T + G + N +
Sbjct: 242 SSSSSSSSSSSSSSSSSDGSISSSLATGESGSDIGLNWI 126
Score = 22.7 bits (47), Expect(2) = 1.1
Identities = 8/9 (88%), Positives = 8/9 (88%)
Frame = -1
Query: 24 PPKPPPLFL 32
PP PPPLFL
Sbjct: 280 PPLPPPLFL 254
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 27.7 bits (60), Expect(2) = 1.6
Identities = 12/17 (70%), Positives = 12/17 (70%)
Frame = -3
Query: 19 PNPLHPPKPPPLFLRRR 35
P PL PP PP L LRRR
Sbjct: 403 PLPLAPPHPPRLHLRRR 353
Score = 20.8 bits (42), Expect(2) = 1.6
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = -2
Query: 46 GFSSSSSSSSSDDVVSTRKST 66
G SSSSSSSSS S+ K +
Sbjct: 263 GASSSSSSSSSSLSSSSSKDS 201
>TC218341 homologue to UP|Q6IEK9 (Q6IEK9) WRKY transcription factor 72,
partial (15%)
Length = 439
Score = 30.0 bits (66), Expect = 1.7
Identities = 26/95 (27%), Positives = 46/95 (48%), Gaps = 8/95 (8%)
Frame = +2
Query: 108 MLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSD 167
+LGL+ +++S+ +VS + + V S GY+ ++ + L + ++AS D
Sbjct: 53 VLGLISMENYSMLFSVSNSSSYPIGVGSSQIGYSGQSSNAFLGLRPSNELAS------DD 214
Query: 168 CEKRSEILEVKGGGE--------DGGEIEVASDLG 194
EKR +GGG+ GG I V+ +LG
Sbjct: 215 HEKR------QGGGDGNMLMSQISGGSINVSDELG 301
>TC228820 weakly similar to UP|Q8LRM1 (Q8LRM1) Nam-like protein 4, partial
(10%)
Length = 616
Score = 29.6 bits (65), Expect = 2.2
Identities = 16/48 (33%), Positives = 24/48 (49%)
Frame = +2
Query: 197 DLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELNARKQEMMQLE 244
D EN SS +PP +I+Q+QSE+ + E + EMM +
Sbjct: 200 DTENLESS------MPPSCAKFIKQMQSEMQKLSVEKETIRFEMMSAQ 325
>TC213408 similar to UP|Q9Y1M2 (Q9Y1M2) P68 RNA helicase, partial (3%)
Length = 802
Score = 29.6 bits (65), Expect = 2.2
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 3/47 (6%)
Frame = +2
Query: 24 PPKPPPLF---LRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTF 67
PP PPP F + + FL SC++ F+ S+ SS + KS+F
Sbjct: 602 PPPPPP*FSL*FQPQESFLQSCHSCPFACSAREKSSFAGSAREKSSF 742
>BM731314
Length = 421
Score = 29.6 bits (65), Expect = 2.2
Identities = 14/30 (46%), Positives = 16/30 (52%)
Frame = +1
Query: 180 GGEDGGEIEVASDLGLKDLENCSSSPRVFG 209
GGE + V DLG L+ C S RVFG
Sbjct: 172 GGEPNSNLHVVEDLGA*KLQTC*LSSRVFG 261
>TC218296
Length = 1248
Score = 29.3 bits (64), Expect = 2.9
Identities = 17/42 (40%), Positives = 23/42 (54%)
Frame = -1
Query: 23 HPPKPPPLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRK 64
H P+ P+F+ +R +T Y GF S SSSS +V T K
Sbjct: 1140 HVPRISPIFISSKRVQITVTY--GFEPSYSSSSWAGLVKTTK 1021
>TC207784 similar to GB|AAM19935.1|20453393|AY097419 AT5g58640/mzn1_90
{Arabidopsis thaliana;} , partial (66%)
Length = 659
Score = 29.3 bits (64), Expect = 2.9
Identities = 21/62 (33%), Positives = 26/62 (41%)
Frame = +1
Query: 8 PFILSRLTDLSPNPLHPPKPPPLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTF 67
P +LS +PNP PL LR + L+S + SSS S ST ST
Sbjct: 70 PVLLSSFNGSNPNPSRWITNLPLLLRHSQPLLSSTLSQTNPSSSPSHPPQTAPSTAPSTT 249
Query: 68 DR 69
R
Sbjct: 250 PR 255
>TC222072
Length = 546
Score = 28.5 bits (62), Expect = 4.9
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = -3
Query: 115 DHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDI 157
+H S+T + + P R + ++ W A +RMSLTR DI
Sbjct: 289 NHISLTXSALEFPSRRTCLKEMLH---CW*ARFRMSLTRGWDI 170
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,589,923
Number of Sequences: 63676
Number of extensions: 265680
Number of successful extensions: 3802
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 2981
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3550
length of query: 360
length of database: 12,639,632
effective HSP length: 98
effective length of query: 262
effective length of database: 6,399,384
effective search space: 1676638608
effective search space used: 1676638608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0081b.1


