

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076c.8
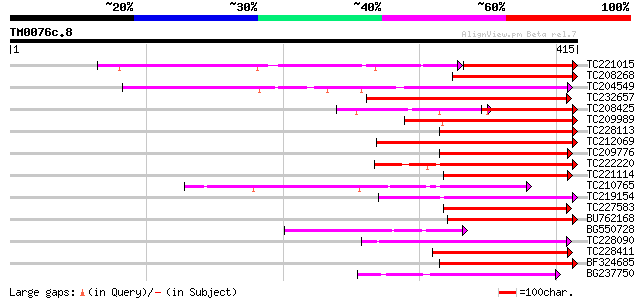
(415 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221015 similar to UP|Q9FNE1 (Q9FNE1) Receptor-like serine/thre... 178 2e-74
TC208268 weakly similar to UP|Q9LMB9 (Q9LMB9) F14D16.24, partial... 174 9e-44
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 171 4e-43
TC232657 weakly similar to UP|O22580 (O22580) Receptor-like seri... 142 4e-34
TC208425 serine/threonine kinase-related protein [Glycine max] 77 1e-27
TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 119 3e-27
TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kin... 111 7e-25
TC212069 weakly similar to UP|Q9M0X5 (Q9M0X5) Receptor protein k... 111 7e-25
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 111 7e-25
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 107 1e-23
TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like prot... 105 4e-23
TC210765 weakly similar to GB|BAC15917.1|22831054|AP003811 recep... 102 3e-22
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 101 7e-22
TC227583 similar to UP|Q9LPQ6 (Q9LPQ6) F15H18.11, partial (14%) 100 9e-22
BU762168 99 3e-21
BG550728 similar to GP|10178148|dbj receptor-like serine/threoni... 97 1e-20
TC228090 similar to UP|Q6QLL5 (Q6QLL5) WAK-like kinase, partial ... 96 2e-20
TC228411 similar to UP|Q94JZ6 (Q94JZ6) Protein kinase-like prote... 96 4e-20
BF324685 weakly similar to GP|20804874|dbj B1100D10.33 {Oryza sa... 94 1e-19
BG237750 93 2e-19
>TC221015 similar to UP|Q9FNE1 (Q9FNE1) Receptor-like serine/threonine
kinase, partial (33%)
Length = 1110
Score = 178 bits (451), Expect(2) = 2e-74
Identities = 101/281 (35%), Positives = 155/281 (54%), Gaps = 14/281 (4%)
Frame = +1
Query: 65 MEKISEQMRTDGFGT---AVAGTGPDTN-YGLAQCYGDLSLLDCVLCYAEARTVLPQCFP 120
ME +S+ + ++ +GT ++G+G YG AQC+ DLS DC+LCYA +RT LP+C P
Sbjct: 34 MESLSQLVTSNNWGTHSVKISGSGSSIPIYGFAQCFRDLSHTDCLLCYAASRTRLPRCLP 213
Query: 121 YNGGRIYLDGCFMRAENYSFFNEYTGPGDRAV-CGNTTRKNSTFQAAAKQAVLSAVQDAP 179
RIYLDGCF+R +NYSF++E T P AV C + + ++ V V +
Sbjct: 214 SVSARIYLDGCFLRYDNYSFYSEGTDPSRDAVNCTGVAAGDEAERVELQERVGRVVDNVV 393
Query: 180 N-----NKGYAKGVVAVSGTANESAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEG 234
N G+ G V E Y LA CW TL S C+ CL A + GCLP EG
Sbjct: 394 NIAERDGNGFGVGEV-------EGVYALAQCWNTLGSGGCRECLRKAGREVKGCLPKKEG 552
Query: 235 RALNTGCFMRYSDTDFLNKEAENGSSGGNVMV----IVVAVVSSVVVTVVGVIIGAYVWK 290
RALN GC++RYS F N++ + G GGN + ++VA V + ++ + +Y
Sbjct: 553 RALNAGCYLRYSTQKFYNEDGDAG--GGNGFLRRRGVIVAEVLAAAAVIMLALSASYAAF 726
Query: 291 RRYIQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATGSFH 331
++ + K+ + + +++ ++ +SLN+KY TL++AT F+
Sbjct: 727 TKFSKIKKENN-NLGQISSSISKSSLNYKYETLEKATDYFN 846
Score = 119 bits (299), Expect(2) = 2e-74
Identities = 54/83 (65%), Positives = 69/83 (83%)
Frame = +2
Query: 333 SNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGC 392
S K+GQGG G+V+KG+L +G+ +A+KRL +NNR +F+NEVN+IS +EHKNLV+LLGC
Sbjct: 851 SRKVGQGGAGSVFKGILPNGKVVAVKRLIFNNRQWVDEFFNEVNLISGIEHKNLVKLLGC 1030
Query: 393 SCSGPESLLVYEFLPNKSLDGFI 415
S GPESLLVYE+LP KSLD FI
Sbjct: 1031SIEGPESLLVYEYLPKKSLDQFI 1099
>TC208268 weakly similar to UP|Q9LMB9 (Q9LMB9) F14D16.24, partial (32%)
Length = 1457
Score = 174 bits (440), Expect = 9e-44
Identities = 82/91 (90%), Positives = 89/91 (97%)
Frame = +2
Query: 325 RATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHK 384
+AT SFHE+NKLGQGGFGTVYKGVLADGREIA+KRL++NNRHRAADFYNEVNIISSVEHK
Sbjct: 2 KATESFHENNKLGQGGFGTVYKGVLADGREIAVKRLFFNNRHRAADFYNEVNIISSVEHK 181
Query: 385 NLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
NLVRLLGCSCSGPESLLVYEFLPN+SLD +I
Sbjct: 182 NLVRLLGCSCSGPESLLVYEFLPNRSLDRYI 274
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 171 bits (434), Expect = 4e-43
Identities = 123/366 (33%), Positives = 174/366 (46%), Gaps = 36/366 (9%)
Frame = +3
Query: 83 GTGP-DTNYGLAQCYGDLSLLDCVLCYAEARTVL-PQCFPYNGGRIYLDGCFMRAENYSF 140
GT P D YGL C GD+ C C A L QC I+ D C +R N SF
Sbjct: 12 GTSPSDRVYGLFMCRGDVPSALCQQCVVNATGRLRSQCSLAKQAVIWYDECTVRYSNRSF 191
Query: 141 FNEY-TGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQDAPNN----KGYAKGVVAVSGTA 195
F+ T P + +F Q + +A N K YA +SG
Sbjct: 192 FSTVDTRPRVGLLNTANISNQDSFMRLLFQTINRTADEAANFSVGLKKYAVNQANISGF- 368
Query: 196 NESAYVLADCWRTLDSSSCKACLENASSSILGCLPW-----SEGRALNTGCFMRYSDTDF 250
+S Y LA C L +C++CL S ++G LPW GR L C +RY F
Sbjct: 369 -QSLYCLAQCTPDLSQENCRSCL----SGVIGDLPWCCQGKQGGRVLYPSCNVRYELYPF 533
Query: 251 LNKEAE-----------------------NGSSGGNVMVIVVAVVSSVVVTVVGVIIGAY 287
A +G S G ++ IVV + +V++ +VG+
Sbjct: 534 YRVTASPPSSSPSPPTLLPPPTSPISPGSSGISAGTIVAIVVPITVAVLIFIVGIC---- 701
Query: 288 VWKRRYIQKKRRGSYDPDKLAKTLQH-NSLNFKYSTLDRATGSFHESNKLGQGGFGTVYK 346
+ R +KK++GS K A + +SL F +ST++ AT F NKLG+GGFG VYK
Sbjct: 702 -FLSRRARKKQQGSVKEGKTAYDIPTVDSLQFDFSTIEAATNKFSADNKLGEGGFGEVYK 878
Query: 347 GVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFL 406
G L+ G+ +A+KRL ++ +F NEV +++ ++H+NLVRLLG G E +LVYE++
Sbjct: 879 GTLSSGQVVAVKRLSKSSGQGGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYV 1058
Query: 407 PNKSLD 412
PNKSLD
Sbjct: 1059PNKSLD 1076
>TC232657 weakly similar to UP|O22580 (O22580) Receptor-like serine/threonine
kinase, partial (35%)
Length = 1208
Score = 142 bits (357), Expect = 4e-34
Identities = 69/150 (46%), Positives = 98/150 (65%)
Frame = +3
Query: 262 GNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLNFKYS 321
G ++ + SS + ++ V++ + R+ + +RR L T+ + LN Y
Sbjct: 6 GKRTLVKILAASSAALALLLVVVTVVFFTRKNVVTRRRERRQFGALLATVNKSKLNMPYE 185
Query: 322 TLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSV 381
L++AT F+E+NKLGQGG G+VYKGV+ DG +AIKRL YN A F+ EVN+IS +
Sbjct: 186 VLEKATNYFNEANKLGQGGSGSVYKGVMPDGNTVAIKRLSYNTTQWAEHFFTEVNLISGI 365
Query: 382 EHKNLVRLLGCSCSGPESLLVYEFLPNKSL 411
HKNLV+LLGCS +GPESLLVYE++PN+SL
Sbjct: 366 HHKNLVKLLGCSITGPESLLVYEYVPNQSL 455
>TC208425 serine/threonine kinase-related protein [Glycine max]
Length = 763
Score = 77.4 bits (189), Expect(2) = 1e-27
Identities = 35/70 (50%), Positives = 50/70 (71%)
Frame = +1
Query: 346 KGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEF 405
KGVL G+EIA+KRL + A +F NE +++ ++H+NLVRLLG G E +L+YE+
Sbjct: 358 KGVLPSGQEIAVKRLSVTSLQGAVEFRNEAALVAKLQHRNLVRLLGFCLEGQEKILIYEY 537
Query: 406 LPNKSLDGFI 415
+PNKSLD F+
Sbjct: 538 IPNKSLDYFL 567
Score = 63.9 bits (154), Expect(2) = 1e-27
Identities = 43/124 (34%), Positives = 64/124 (50%), Gaps = 10/124 (8%)
Frame = +2
Query: 240 GCFMRYSDTDFLN----KEAENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQ 295
GC +RY F N + SSG + + I++A+V + V ++ I+G Y ++R
Sbjct: 20 GCSVRYELFPFYNVSTVSRLPSPSSGKSSISIILAIVVPITVAILLFIVGVYFLRKR--A 193
Query: 296 KKRRGSYDPDKLAKTLQH----NSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGV--L 349
K+ ++ D +A L SL F T++ AT F + NK+GQGGFG YK V L
Sbjct: 194 SKKYNTFVQDSIADDLTDVGDVESLQFDLPTVEAATNRFSDENKIGQGGFGXGYKRVSSL 373
Query: 350 ADGR 353
AD R
Sbjct: 374 ADRR 385
>TC209989 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (32%)
Length = 738
Score = 119 bits (298), Expect = 3e-27
Identities = 61/128 (47%), Positives = 87/128 (67%), Gaps = 2/128 (1%)
Frame = +3
Query: 290 KRRYIQKKRRGSYDPDKLAKTLQHNS--LNFKYSTLDRATGSFHESNKLGQGGFGTVYKG 347
+ + IQ + G D +LA ++ + L F+++T+ AT +F ++NKLGQGGFG VYKG
Sbjct: 9 RNKPIQNEGEGDDDEGELANDIKTDDQLLQFEFATIKFATNNFSDANKLGQGGFGIVYKG 188
Query: 348 VLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLP 407
L+DG+EIAIKRL N+ +F NE+ + ++H+NLVRLLG + E LL+YEF+P
Sbjct: 189 TLSDGQEIAIKRLSINSNQGETEFKNEILLTGRLQHRNLVRLLGFCFARRERLLIYEFVP 368
Query: 408 NKSLDGFI 415
NKSLD FI
Sbjct: 369 NKSLDYFI 392
>TC228113 similar to UP|Q9XED4 (Q9XED4) Receptor-like protein kinase homolog
RK20-1, partial (53%)
Length = 1492
Score = 111 bits (277), Expect = 7e-25
Identities = 54/101 (53%), Positives = 72/101 (70%)
Frame = +1
Query: 315 SLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNE 374
SL F + T+ AT F +SNKLGQGGFG VY+G L+DG+ IA+KRL + +F NE
Sbjct: 109 SLQFNFDTIRVATEDFSDSNKLGQGGFGAVYRGRLSDGQMIAVKRLSRESSQGDTEFKNE 288
Query: 375 VNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
V +++ ++H+NLVRLLG G E LL+YE++PNKSLD FI
Sbjct: 289 VLLVAKLQHRNLVRLLGFCLEGKERLLIYEYVPNKSLDYFI 411
>TC212069 weakly similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like
protein, partial (19%)
Length = 513
Score = 111 bits (277), Expect = 7e-25
Identities = 55/147 (37%), Positives = 91/147 (61%)
Frame = +1
Query: 269 VAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATG 328
+ +V +VV++V + +G + R K + + + +L F+ + ++ AT
Sbjct: 1 IVIVVPIVVSLVLLSLGCCCFLHRKATKNQHDILKENFGNDSTTLETLRFELAKIEAATN 180
Query: 329 SFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVR 388
F + N +G+GGFG VY+G+L DG+EIA+KRL ++R A +F NEV +I+ ++H+NLVR
Sbjct: 181 RFAKENMIGKGGFGEVYRGILLDGQEIAVKRLTGSSRQGAVEFKNEVQVIAKLQHRNLVR 360
Query: 389 LLGCSCSGPESLLVYEFLPNKSLDGFI 415
L G E +L+YE++PNKSLD F+
Sbjct: 361 LQGFCLEDDEKILIYEYVPNKSLDYFL 441
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 111 bits (277), Expect = 7e-25
Identities = 53/98 (54%), Positives = 71/98 (72%)
Frame = +2
Query: 315 SLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNE 374
SL F +ST++ AT F E+NKLG+GGFG VYKG+L G+E+A+KRL + +F NE
Sbjct: 35 SLRFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNE 214
Query: 375 VNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
V I++ ++H+NLVRLLG G E +LVYEF+ NKSLD
Sbjct: 215 VEIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLD 328
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 107 bits (267), Expect = 1e-23
Identities = 62/154 (40%), Positives = 94/154 (60%), Gaps = 6/154 (3%)
Frame = +3
Query: 268 VVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDP------DKLAKTLQHNSLNFKYS 321
VV +V VVV+++ ++ Y +I K+ R Y+ + + TL+ SL F
Sbjct: 42 VVLIVVPVVVSIILLLCVCY-----FILKRSRKKYNTLLRENFGEESATLE--SLQFGLV 200
Query: 322 TLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSV 381
T++ AT F ++G+GGFG VYKGVL DGREIA+K+L ++ A +F NE+ +I+ +
Sbjct: 201 TIEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIAKL 380
Query: 382 EHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
+H+NLV LLG E +L+YEF+ NKSLD F+
Sbjct: 381 QHRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFL 482
>TC221114 similar to UP|Q8RZ15 (Q8RZ15) Receptor kinase-like protein, partial
(29%)
Length = 946
Score = 105 bits (262), Expect = 4e-23
Identities = 50/95 (52%), Positives = 66/95 (68%)
Frame = +3
Query: 318 FKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNI 377
F + + ATG+F +NKLGQGGFG VYKGVL DG+EIAIKRL + +F NE +
Sbjct: 408 FSFPIIAAATGNFSVANKLGQGGFGPVYKGVLPDGQEIAIKRLSSRSGQGLVEFKNEAEL 587
Query: 378 ISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
++ ++H NLVRL G E++L+YE+LPNKSLD
Sbjct: 588 VAKLQHTNLVRLSGLCIQNEENILIYEYLPNKSLD 692
>TC210765 weakly similar to GB|BAC15917.1|22831054|AP003811 receptor
kinase-like protein {Oryza sativa (japonica
cultivar-group);} , partial (10%)
Length = 787
Score = 102 bits (254), Expect = 3e-22
Identities = 81/269 (30%), Positives = 136/269 (50%), Gaps = 15/269 (5%)
Frame = +1
Query: 129 DGCFMRAENYSFFNEYTGPGDRAVCGNTTRKNSTFQAAAKQAVLSAVQD-------APNN 181
D C + N+ F TG RA+ + +++ + + + S + D + +
Sbjct: 1 DECTLYFTNHYFSR--TGIEPRAMLSDDRNISASDLDSFNRTLFSLLNDLAEEAANSQSA 174
Query: 182 KGYAKGVVAVSGTANE-SAYVLADCWRTLDSSSCKACLENASSSILGCLPWSEG-RALNT 239
K +A G G++ + + Y LA+C T S+ C+ CL+NA S++ C +G RAL
Sbjct: 175 KKFATGQSRFDGSSPQRTVYALAECAPTETSTQCEECLKNAISTLPSCCGGKQGARALLA 354
Query: 240 GCFMRYSDTDFLNKE------AENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRY 293
C +RY F N A N S V++IVV VV SV + + GV R+
Sbjct: 355 HCDVRYELFLFYNTSGTSVIYAGNKKSVSRVILIVVPVVVSVFL-LCGVCYFILKRSRKR 531
Query: 294 IQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGR 353
+ R ++ + + TL+ +L F +T++ AT F ++G+GGFG VYKG+ DGR
Sbjct: 532 YKTLLRENFGEE--SATLE--ALQFGLATIEAATNKFSYERRIGEGGFGVVYKGIFPDGR 699
Query: 354 EIAIKRLYYNNRHRAADFYNEVNIISSVE 382
EIA+K+L ++ A +F NE+ +I+ ++
Sbjct: 700 EIAVKKLSRSSGQGAIEFKNEILLIAKLQ 786
Score = 32.3 bits (72), Expect = 0.40
Identities = 21/62 (33%), Positives = 28/62 (44%), Gaps = 2/62 (3%)
Frame = +1
Query: 83 GTGPD-TNYGLAQCYGDLSLLDCVLCYAEARTVLPQCF-PYNGGRIYLDGCFMRAENYSF 140
G+ P T Y LA+C + C C A + LP C G R L C +R E + F
Sbjct: 208 GSSPQRTVYALAECAPTETSTQCEECLKNAISTLPSCCGGKQGARALLAHCDVRYELFLF 387
Query: 141 FN 142
+N
Sbjct: 388 YN 393
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 101 bits (251), Expect = 7e-22
Identities = 56/145 (38%), Positives = 84/145 (57%)
Frame = +1
Query: 271 VVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLNFKYSTLDRATGSF 330
++ + V +GV++ RR I K + D+ + + F T+ AT +F
Sbjct: 178 IIGTSVAAPLGVVLAICFIYRRNIADKSKTKKSIDRQLQDVDVPL--FDMLTITAATDNF 351
Query: 331 HESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISSVEHKNLVRLL 390
+NK+G+GGFG VYKG L G+EIA+KRL + +F EV +I+ ++H+NLV+LL
Sbjct: 352 LLNNKIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQHRNLVKLL 531
Query: 391 GCSCSGPESLLVYEFLPNKSLDGFI 415
GC G E LLVYE++ N SL+ FI
Sbjct: 532 GCCIKGQEKLLVYEYVVNGSLNSFI 606
>TC227583 similar to UP|Q9LPQ6 (Q9LPQ6) F15H18.11, partial (14%)
Length = 838
Score = 100 bits (250), Expect = 9e-22
Identities = 50/95 (52%), Positives = 64/95 (66%), Gaps = 1/95 (1%)
Frame = +1
Query: 318 FKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNI 377
F Y L+ AT F S +LG+GGFGTVY G L DGR +A+KRLY NN R A F NE+ I
Sbjct: 40 FLYDELEEATNYFDSSKELGEGGFGTVYFGKLRDGRSVAVKRLYENNFKRVAQFMNEIKI 219
Query: 378 ISSVEHKNLVRLLGC-SCSGPESLLVYEFLPNKSL 411
++ ++H NLV L GC S E LLVYE++PN ++
Sbjct: 220 LAKLDHPNLVTLFGCTSRHTRELLLVYEYIPNGTI 324
>BU762168
Length = 341
Score = 99.4 bits (246), Expect = 3e-21
Identities = 49/95 (51%), Positives = 65/95 (67%)
Frame = +1
Query: 321 STLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVNIISS 380
S + AT F E NK+G+GGFG+VY G L G EIA+KRL N+ ++F NEV +I+
Sbjct: 55 SIIIAATNKFSEGNKIGEGGFGSVYWGKLPSGLEIAVKRLSKNSDQGMSEFVNEVKLIAK 234
Query: 381 VEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
V+H+NLV+LLGC E +LVYE++ N SLD FI
Sbjct: 235 VQHRNLVKLLGCCIKKQEIMLVYEYMVNGSLDYFI 339
>BG550728 similar to GP|10178148|dbj receptor-like serine/threonine kinase
{Arabidopsis thaliana}, partial (8%)
Length = 395
Score = 97.4 bits (241), Expect = 1e-20
Identities = 49/134 (36%), Positives = 78/134 (57%)
Frame = +3
Query: 202 LADCWRTLDSSSCKACLENASSSILGCLPWSEGRALNTGCFMRYSDTDFLNKEAENGSSG 261
LA CW+T+ C CL A + + GCLP EGRALNTGC++RYS F N+ ++G
Sbjct: 3 LAQCWKTVGVKGCSDCLRKAENEVKGCLPKREGRALNTGCYLRYSTVKFYNQGGQDGQGD 182
Query: 262 GNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLNFKYS 321
+ V+ SV+ V V++ V + +K+R+ ++ ++ +L+++SLN+KY
Sbjct: 183 DSSRKRVIIAAGSVLAAAV-VVLTLAVSYVAFTKKRRKNNF--IEVPPSLKNSSLNYKYE 353
Query: 322 TLDRATGSFHESNK 335
TL++AT F S K
Sbjct: 354 TLEKATDYFSSSRK 395
Score = 38.9 bits (89), Expect = 0.004
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 2/60 (3%)
Frame = +3
Query: 92 LAQCYGDLSLLDCVLCYAEARTVLPQCFPYNGGRIYLDGCFMRAENYSFFNE--YTGPGD 149
LAQC+ + + C C +A + C P GR GC++R F+N+ G GD
Sbjct: 3 LAQCWKTVGVKGCSDCLRKAENEVKGCLPKREGRALNTGCYLRYSTVKFYNQGGQDGQGD 182
>TC228090 similar to UP|Q6QLL5 (Q6QLL5) WAK-like kinase, partial (51%)
Length = 1533
Score = 96.3 bits (238), Expect = 2e-20
Identities = 58/155 (37%), Positives = 84/155 (53%), Gaps = 1/155 (0%)
Frame = +1
Query: 258 GSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHNSLN 317
G GG IV+ + VV + V +G+ R K R + +L + NS+
Sbjct: 1 GRCGGTTRFIVL-IGGFVVGVSLMVTLGSLCCFYRRRSKLRVTNSTKRRLTEATGKNSVP 177
Query: 318 -FKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNEVN 376
+ Y +++AT SF E +LG G +GTVY G L + +AIKR+ + + NE+
Sbjct: 178 IYPYKDIEKATNSFSEKQRLGTGAYGTVYAGKLYNNEWVAIKRIKHRDTDSIEQVMNEIK 357
Query: 377 IISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSL 411
++SSV H NLVRLLGCS E +LVYEF+PN +L
Sbjct: 358 LLSSVSHTNLVRLLGCSIEYGEQILVYEFMPNGTL 462
>TC228411 similar to UP|Q94JZ6 (Q94JZ6) Protein kinase-like protein
(At3g24600), partial (28%)
Length = 751
Score = 95.5 bits (236), Expect = 4e-20
Identities = 47/103 (45%), Positives = 68/103 (65%)
Frame = +3
Query: 310 TLQHNSLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAA 369
+L + F Y L RAT F ++N LGQGGFG V++G+L +G+E+A+K+L +
Sbjct: 9 SLGFSKSTFTYEELARATDGFSDANLLGQGGFGYVHRGILPNGKEVAVKQLKAGSGQGER 188
Query: 370 DFYNEVNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLD 412
+F EV IIS V HK+LV L+G +G + LLVYEF+PN +L+
Sbjct: 189 EFQAEVEIISRVHHKHLVSLVGYCITGSQRLLVYEFVPNNTLE 317
>BF324685 weakly similar to GP|20804874|dbj B1100D10.33 {Oryza sativa
(japonica cultivar-group)}, partial (11%)
Length = 361
Score = 93.6 bits (231), Expect = 1e-19
Identities = 46/101 (45%), Positives = 64/101 (62%)
Frame = +3
Query: 315 SLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNE 374
SL F +T++ AT F + NK+GQGGFG VYKGV EI +KRL A F NE
Sbjct: 3 SLQFDLATVEAATDRFSDENKIGQGGFGVVYKGVXXXXXEITVKRLLVTFLLGAV*FRNE 182
Query: 375 VNIISSVEHKNLVRLLGCSCSGPESLLVYEFLPNKSLDGFI 415
+++ + H +L+RL G G E++L+YE++PNKSLD F+
Sbjct: 183 PVLVAKLHHTSLMRLWGFCLEGHETILIYEYIPNKSLDRFL 305
>BG237750
Length = 428
Score = 93.2 bits (230), Expect = 2e-19
Identities = 52/149 (34%), Positives = 89/149 (58%)
Frame = +1
Query: 255 AENGSSGGNVMVIVVAVVSSVVVTVVGVIIGAYVWKRRYIQKKRRGSYDPDKLAKTLQHN 314
+E+GSS +++ +VVAVV V + ++I +W +R++ +R + + K +
Sbjct: 1 SESGSSSISIIRVVVAVV---VAGAIIILIFGILWWKRFLGWERSVARE----LKGFESQ 159
Query: 315 SLNFKYSTLDRATGSFHESNKLGQGGFGTVYKGVLADGREIAIKRLYYNNRHRAADFYNE 374
+ F + AT +F +S K+G+GGFG VYKGVL+DG A+K+L +R + +F NE
Sbjct: 160 TSLFTLRQIKAATNNFDKSLKIGEGGFGLVYKGVLSDGTVXAVKQLSTRSRQGSREFVNE 339
Query: 375 VNIISSVEHKNLVRLLGCSCSGPESLLVY 403
+ +IS+++H LV+L GC + LL+Y
Sbjct: 340 IGLISALQHPCLVKLYGCCMEEDQLLLIY 426
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,151,662
Number of Sequences: 63676
Number of extensions: 278485
Number of successful extensions: 1947
Number of sequences better than 10.0: 408
Number of HSP's better than 10.0 without gapping: 1819
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1839
length of query: 415
length of database: 12,639,632
effective HSP length: 100
effective length of query: 315
effective length of database: 6,272,032
effective search space: 1975690080
effective search space used: 1975690080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0076c.8


