

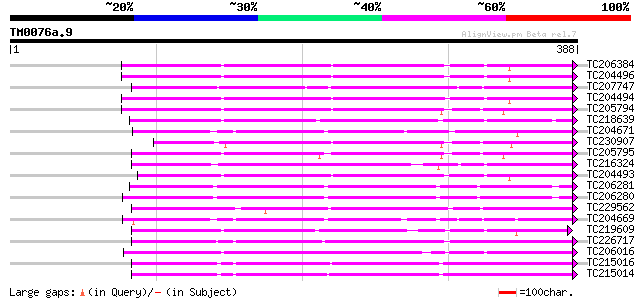
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0076a.9
(388 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC206384 UP|O23961 (O23961) Peroxidase precursor, complete 227 8e-60
TC204496 homologue to UP|O23961 (O23961) Peroxidase precursor, p... 226 1e-59
TC207747 weakly similar to UP|Q9M4Z5 (Q9M4Z5) Peroxidase prx12 p... 216 2e-56
TC204494 similar to UP|O23961 (O23961) Peroxidase precursor, par... 213 2e-55
TC205794 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), ... 207 6e-54
TC218639 similar to UP|PE17_ARATH (Q9SJZ2) Peroxidase 17 precurs... 200 1e-51
TC204671 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2... 196 1e-50
TC230907 similar to UP|O24081 (O24081) Peroxidase1A precursor ,... 191 5e-49
TC205795 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), ... 191 6e-49
TC216324 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, complete 189 2e-48
TC204493 UP|O22443 (O22443) Seed coat peroxidase precursor , co... 186 2e-47
TC206281 UP|Q9ZRG5 (Q9ZRG5) Peroxidase (Fragment) , complete 186 2e-47
TC206280 UP|Q9ZRG6 (Q9ZRG6) Peroxidase (Fragment) , complete 183 1e-46
TC229562 similar to GB|CAA70034.1|1620369|ATP22A peroxidase ATP2... 183 1e-46
TC204669 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2... 181 5e-46
TC219609 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1... 181 7e-46
TC226717 homologue to UP|Q9ZTW8 (Q9ZTW8) Peroxidase, partial (85%) 181 7e-46
TC206016 similar to UP|Q6T1C8 (Q6T1C8) Peroxidase precursor, par... 181 7e-46
TC215016 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (... 180 1e-45
TC215014 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (... 180 1e-45
>TC206384 UP|O23961 (O23961) Peroxidase precursor, complete
Length = 1251
Score = 227 bits (578), Expect = 8e-60
Identities = 124/315 (39%), Positives = 184/315 (58%), Gaps = 3/315 (0%)
Frame = +1
Query: 77 PNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASIL 136
P +++ L FY D+CP I+ + + + +P+ +A+++RL FHDCFV GCDAS+L
Sbjct: 88 PLSLDAQLDPSFYRDTCPRVHSIVREVVRNVSKKDPRMLASLIRLHFHDCFVQGCDASVL 267
Query: 137 LDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAG 196
L++T ++ E+ + N L+G D+V+DIK+ +E+ CPG+VSCAD + S + L G
Sbjct: 268 LNNTATIES-EQQALPNNNSLRGLDVVNDIKTAVEKACPGVVSCADILTLASEISSVLGG 444
Query: 197 MPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIG 256
P K GRRDSL + + + N LP P ++ ++ F +G ++V L GAH+ G
Sbjct: 445 GPHWKVPLGRRDSLTANRNLANQN-LPAPFFNLSRLKAAFAVQGLDTTDLVALSGAHTFG 621
Query: 257 AAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDN-TPTVMD 315
AHC+ +DR+YNF T KPDP L +L +LRQIC N G N VNFD TP +D
Sbjct: 622 RAHCNFILDRLYNFSGTGKPDPTLDTTYLQQLRQICPNGGP----NNLVNFDPVTPDKID 789
Query: 316 NLFYRDLVDKGKSLLLTDAHLVTDP--RTAPTVGQMADDQALFHKRFAEVMTKLTSLNVL 373
+++ +L K K LL +D L + P T P V + + DQ +F F M K+ ++ VL
Sbjct: 790 RVYFSNLQVK-KGLLQSDQELFSTPGADTIPIVNRFSSDQKVFFDAFEASMIKMGNIGVL 966
Query: 374 TGNDGEVRKICRATN 388
TG GE+RK C N
Sbjct: 967 TGKKGEIRKHCNFVN 1011
>TC204496 homologue to UP|O23961 (O23961) Peroxidase precursor, partial (97%)
Length = 2193
Score = 226 bits (576), Expect = 1e-59
Identities = 125/315 (39%), Positives = 184/315 (57%), Gaps = 3/315 (0%)
Frame = +3
Query: 77 PNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASIL 136
P +++ L FY D+CP I+ + + + +P+ +A+++RL FHDCFV GCDAS+L
Sbjct: 927 PLSLDAQLDPSFYRDTCPKVHSIVREVVRNVSKKDPRMLASLIRLHFHDCFVQGCDASVL 1106
Query: 137 LDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAG 196
L++T ++ E+ + N L+G D+V+DIK+ +E+ CPG+VSCAD + S + L G
Sbjct: 1107 LNNTATIES-EQQALPNNNSLRGLDVVNDIKTAVEQACPGVVSCADILTLASEISSILGG 1283
Query: 197 MPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIG 256
P K GRRDSL + T+ + N LP P ++ ++ F +G ++V L GAH+ G
Sbjct: 1284 GPDWKVPLGRRDSLTANRTLANQN-LPAPFFNLTQLKAAFAVQGLDTTDLVALSGAHTFG 1460
Query: 257 AAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDN-TPTVMD 315
AHC + R+YNF T KPDP L +L +LRQIC N G N VNFD TP +D
Sbjct: 1461 RAHCSFILGRLYNFSGTGKPDPTLDTTYLQQLRQICPNGGP----NNLVNFDPVTPDKID 1628
Query: 316 NLFYRDLVDKGKSLLLTDAHLVTDP--RTAPTVGQMADDQALFHKRFAEVMTKLTSLNVL 373
+++ +L K K LL +D L + P T P V + + DQ +F F M K+ ++ VL
Sbjct: 1629 RVYFSNLQVK-KGLLQSDQELFSTPGADTIPIVNRFSSDQNVFFDAFEASMIKMGNIGVL 1805
Query: 374 TGNDGEVRKICRATN 388
TGN GE+RK C N
Sbjct: 1806 TGNKGEIRKHCNFVN 1850
>TC207747 weakly similar to UP|Q9M4Z5 (Q9M4Z5) Peroxidase prx12 precursor ,
partial (88%)
Length = 1204
Score = 216 bits (549), Expect = 2e-56
Identities = 124/306 (40%), Positives = 182/306 (58%), Gaps = 1/306 (0%)
Frame = +3
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ G+YS SC AE I+ D + V NP A ++R+ FHDCF+ GCDAS+LLDSTP
Sbjct: 123 LQVGYYSYSCSMAEFIVKDEVRKGVTNNPGIAAGLVRMHFHDCFIRGCDASVLLDSTPL- 299
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALA-GMPRQKP 202
+ EK S N L+G +++D+ K+KLE CPGIVSCAD + F + +++ A G+ P
Sbjct: 300 NTAEKDSPANKPSLRGYEVIDNAKAKLEAVCPGIVSCADIVAFAARDSVEFARGLGYDVP 479
Query: 203 LGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
GRRD SLA+ LP P ++ +++ +LF RKG + +EMV L GAH+IG +HC
Sbjct: 480 -AGRRDGRISLASDTR-TELPPPTFNVNQLTQLFARKGLTQDEMVTLSGAHTIGRSHCSA 653
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDL 322
F R+YNF T+ DP+L P + L++ C T + P++ ++P + D +Y D+
Sbjct: 654 FSSRLYNFSTTSSQDPSLDPSYAALLKRQCPQGSTNQNLVVPMD-PSSPGIADVGYYVDI 830
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRK 382
+ + L +D L+T+ TA V Q A D L+ +FA+ M K+ + VL GN GE+R
Sbjct: 831 L-ANRGLFTSDQTLLTNAETASQVKQNARDPYLWASQFADAMVKMGQIIVLKGNAGEIRT 1007
Query: 383 ICRATN 388
CR N
Sbjct: 1008NCRVVN 1025
>TC204494 similar to UP|O23961 (O23961) Peroxidase precursor, partial (93%)
Length = 1326
Score = 213 bits (541), Expect = 2e-55
Identities = 125/315 (39%), Positives = 178/315 (55%), Gaps = 3/315 (0%)
Frame = +3
Query: 77 PNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASIL 136
P + N L FY +CP I+ ++ RT+P+ A+++RL FHDCFV GCDASIL
Sbjct: 132 PFSSNAKLEPCFYKKTCPQVHFIVFKVVEKVSRTDPRMPASLVRLFFHDCFVQGCDASIL 311
Query: 137 LDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAG 196
L++T + E+ + N ++G D+V+ IK++LE+ CPG+VSCAD + + + LA
Sbjct: 312 LNNTATIVS-EQQALPNNNSIRGLDVVNQIKTELEKACPGVVSCADILTLAAEVSSVLAH 488
Query: 197 MPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIG 256
P K GRRDSL + T+ + N LP P ++ ++ F +G ++V L GAHS G
Sbjct: 489 GPYLKFPLGRRDSLTANRTLANQN-LPAPFFNLTQLKAAFAVQGLDTTDLVALSGAHSFG 665
Query: 257 AAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMD 315
C +DR+YNF T +PDP L +L +LRQIC G P N VNFD TP +D
Sbjct: 666 RVRCLFILDRLYNFSGTGRPDPTLDTTYLKQLRQICPQGGPP---NNLVNFDPTTPDTLD 836
Query: 316 NLFYRDLVDKGKSLLLTDAHLVTDP--RTAPTVGQMADDQALFHKRFAEVMTKLTSLNVL 373
+Y +L K K LL +D L + P T V + + DQ F K F+ M K+ ++ VL
Sbjct: 837 KNYYSNLQVK-KGLLQSDQELFSTPGADTISIVNKFSSDQIAFFKSFSASMIKMGNIGVL 1013
Query: 374 TGNDGEVRKICRATN 388
TG GE+RK C N
Sbjct: 1014TGKKGEIRKQCNFVN 1058
>TC205794 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1205
Score = 207 bits (527), Expect = 6e-54
Identities = 119/317 (37%), Positives = 179/317 (55%), Gaps = 5/317 (1%)
Frame = +2
Query: 77 PNTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASIL 136
P + + L FY D+CP I+ + + +++P+ +A+++RL FHDCFV GCDASIL
Sbjct: 95 PFSSDAQLDPSFYRDTCPKVHSIVREVVRNVSKSDPRMLASLIRLHFHDCFVQGCDASIL 274
Query: 137 LDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAG 196
L++T ++ E+ +F N ++G D+V+ IK+ +E CPG+VSCAD + + + L
Sbjct: 275 LNNTATIES-EQQAFPNNNSIRGLDVVNQIKTAVENACPGVVSCADILALAAEISSVLGH 451
Query: 197 MPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIG 256
P K GRRDSL + T+ + N LP P + ++ + F +G + ++V L GAH+IG
Sbjct: 452 GPDWKVPLGRRDSLTANRTLANQN-LPAPFLNLTQLKDAFAVQGLNTTDLVALSGAHTIG 628
Query: 257 AAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSN--PGTPRYRNEPVNFD-NTPTV 313
A C F+DR+YNF +T PDP L +L L IC N PGT NFD TP
Sbjct: 629 RAQCRFFVDRLYNFSSTGNPDPTLNTTYLQTLSAICPNGGPGT-----NLTNFDPTTPDT 793
Query: 314 MDNLFYRDLVDKGKSLLLTDAHL--VTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLN 371
+D+ +Y +L K LL +D L T T V + +Q LF + F M K+ ++
Sbjct: 794 VDSNYYSNL-QVNKGLLQSDQELFSTTGTDTIAIVNSFSSNQTLFFENFKASMIKMGNIG 970
Query: 372 VLTGNDGEVRKICRATN 388
VLTG+ GE+R+ C N
Sbjct: 971 VLTGSQGEIRQQCNFIN 1021
>TC218639 similar to UP|PE17_ARATH (Q9SJZ2) Peroxidase 17 precursor (Atperox
P17) (ATP25a) , partial (93%)
Length = 1085
Score = 200 bits (508), Expect = 1e-51
Identities = 118/307 (38%), Positives = 171/307 (55%), Gaps = 1/307 (0%)
Frame = +2
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
+LR GFYS +CP AE I+ D + + +++A+++R QFHDCFV GCD S+LLD T
Sbjct: 56 DLRAGFYSKTCPKAEVIVRDVMKKALMREARSVASVMRFQFHDCFVNGCDGSMLLDDTAT 235
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKP 202
EK + N L+ +VD +K LE++CPG+VSCAD ++ S +A+AL G P +
Sbjct: 236 MLG-EKMALSNINSLRSYKVVDQVKQALEKDCPGVVSCADIIIMASRDAVALTGGPEWEV 412
Query: 203 LGGRRDSLYSLATVVDDNN-LPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCD 261
GR DSL A D NN +P P +A +++LFQ+ + +++V L G+HSIG C
Sbjct: 413 RLGRLDSL--SANQEDSNNIMPSPRANASSLIDLFQKYNLTVKDLVALSGSHSIGQGRCF 586
Query: 262 IFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRD 321
M R+YN T +PDPA+ P + L ++C +N N D+TP V DN + +D
Sbjct: 587 SVMFRLYNQSGTGRPDPAIDPSYRQYLNRLCP---LDVDQNVTGNLDSTPLVFDNQYIKD 757
Query: 322 LVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVR 381
L + + L +D L T P T V + + F K + E M K+ L +G GEVR
Sbjct: 758 LAAR-RGFLNSDQTLFTFPHTREFVRLFSRRKTEFFKAYVEGMLKMGDLQ--SGRPGEVR 928
Query: 382 KICRATN 388
CR N
Sbjct: 929 TNCRLVN 949
>TC204671 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (90%)
Length = 1185
Score = 196 bits (498), Expect = 1e-50
Identities = 116/309 (37%), Positives = 172/309 (55%), Gaps = 5/309 (1%)
Frame = +2
Query: 85 REGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGD 144
R GFYS +CP AE I+ V ++ A +LR+ FHDCFV GCDAS+L+ G
Sbjct: 152 RVGFYSSTCPRAESIVKSTVTTHVNSDSTLAAGLLRMHFHDCFVQGCDASVLI----AGS 319
Query: 145 NVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLG 204
E+++F N + L+G +++DD K +LE CPG+VSCAD + + +++ L+G + L
Sbjct: 320 GTERTAFAN-LGLRGFEVIDDAKKQLEAACPGVVSCADILALAARDSVVLSGGLSYQVLT 496
Query: 205 GRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIFM 264
GRRD S A+ D +NLP P S D + F KG + +++V L+GAH+IG C F
Sbjct: 497 GRRDGRISQAS--DVSNLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGAHTIGTTACQFFS 670
Query: 265 DRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDLVD 324
+R+YNF N PDP++ P FL++L+ +C G R V D +L Y +
Sbjct: 671 NRLYNF-TANGPDPSIDPSFLSQLQSLCPQNGDGSKR---VALDTGSQTKFDLSYYSNLR 838
Query: 325 KGKSLLLTDAHLVTDPRTAPTV----GQMADDQAL-FHKRFAEVMTKLTSLNVLTGNDGE 379
+ +L +D L +D T TV G + L F+ F + M K+ ++ + TG DGE
Sbjct: 839 NSRGILQSDQALWSDASTKTTVQRYLGLIRGLLGLTFNVEFGKSMVKMGNIELKTGTDGE 1018
Query: 380 VRKICRATN 388
+RKIC A N
Sbjct: 1019IRKICSAIN 1045
>TC230907 similar to UP|O24081 (O24081) Peroxidase1A precursor , partial
(88%)
Length = 944
Score = 191 bits (485), Expect = 5e-49
Identities = 113/297 (38%), Positives = 169/297 (56%), Gaps = 7/297 (2%)
Frame = +2
Query: 99 IIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNV--EKSSFFNGIL 156
I+ + + + +++P+ +A+++RL FHDCFV GCDASILL+ T D + E+S+ N
Sbjct: 5 IVREVLSNVSQSDPRILASLIRLHFHDCFVQGCDASILLNDT---DTIVSEQSAVPNNNS 175
Query: 157 LKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRRDSLYSLATV 216
++G D+V+ IK+ +E CPGIVSCAD + + + LA P + GRRDSL + T+
Sbjct: 176 IRGLDVVNQIKTAVENACPGIVSCADILALAAQISSDLANGPVWQVPLGRRDSLTANQTL 355
Query: 217 VDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIFMDRVYNFKNTNKP 276
+ N LP P ++ D+++E F + + ++V L GAH+IG A C F+DR+YNF NT P
Sbjct: 356 ANQN-LPAPTFTIDQLIESFGNQSLNITDLVALSGAHTIGRAQCRFFVDRLYNFSNTGNP 532
Query: 277 DPALRPPFLNELRQICSN--PGTPRYRNEPVNFD-NTPTVMDNLFYRDLVDKGKSLLLTD 333
DP L L L+ IC N PGT N D TP D+ +Y +L LL +D
Sbjct: 533 DPTLNTTLLQSLQGICPNGGPGT-----NLTNLDLTTPDTFDSNYYSNL-QLQNGLLQSD 694
Query: 334 AHLVTDPRT--APTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRKICRATN 388
L++ T V +Q LF + F M K+ ++ VLTG+ GE+R C + N
Sbjct: 695 QELLSANNTDIVAIVNNFISNQTLFFENFKASMIKMGNIGVLTGSQGEIRSQCNSVN 865
>TC205795 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(96%)
Length = 1321
Score = 191 bits (484), Expect = 6e-49
Identities = 118/313 (37%), Positives = 168/313 (52%), Gaps = 8/313 (2%)
Frame = +2
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L FY D+CP I+ + + +++P+ +A+++RL FHDCFV GCDASILL+ T
Sbjct: 116 LDNSFYKDTCPRVHSIVREVVRNVSKSDPRILASLIRLHFHDCFVQGCDASILLNDTATI 295
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
+ E+S+ N ++G D+V+ IK+ +E CPGIVSCAD + + + LA P K
Sbjct: 296 VS-EQSAPPNNNSIRGLDVVNQIKTAVENACPGIVSCADILALAAEISSVLAHGPDWKVP 472
Query: 204 GGRRDSL---YSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHC 260
GRRDSL +SLA NLP N++ D++ F R+G + ++V L GAH+IG + C
Sbjct: 473 LGRRDSLNSSFSLAL----QNLPGFNFTLDQLKSTFDRQGLNTTDLVALSGAHTIGRSQC 640
Query: 261 DIFMDRVYNFKNTNKPDPALRPPFLNELRQICSN--PGTPRYRNEPVNFD-NTPTVMDNL 317
F R+YNF DP L LR IC N PGT N D TP D+
Sbjct: 641 RFFAHRIYNFSGNGNSDPTLNTTLSQALRAICPNGGPGT-----NLTNLDLTTPDRFDSN 805
Query: 318 FYRDLVDKGKSLLLTDAHL--VTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTG 375
+Y +L LL +D L + T V +Q LF++ F M K++ + VLTG
Sbjct: 806 YYSNL-QLQNGLLRSDQVLFSTSGAETIAIVNSFGSNQTLFYEHFKVSMIKMSIIEVLTG 982
Query: 376 NDGEVRKICRATN 388
+ GE+RK C N
Sbjct: 983 SQGEIRKHCNFVN 1021
>TC216324 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, complete
Length = 1246
Score = 189 bits (480), Expect = 2e-48
Identities = 111/308 (36%), Positives = 168/308 (54%), Gaps = 3/308 (0%)
Frame = +2
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L E FY CP + + P+ A+I+RL FHDCFV GCD S+LLD G
Sbjct: 128 LSENFYDSKCPKVFYAVKSVLQSALAKEPRQGASIVRLFFHDCFVNGCDGSVLLD----G 295
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
+ EK++ N L+G +++D IKSK+E CPG+VSCAD + + +++A+ G P K
Sbjct: 296 PSSEKTAPPNNNSLRGYEVIDAIKSKVETVCPGVVSCADIVTIAARDSVAILGGPNWKVK 475
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GRRDS + + LP PN S +++ F +G S ++MV L GAH+IG A C +
Sbjct: 476 LGRRDSTTGFFNLANSGVLPGPNSSLSSLIQRFDDQGLSTKDMVALSGAHTIGKARCVSY 655
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQIC--SNPGTPRYRN-EPVNFDNTPTVMDNLFYR 320
DR+YN N + F ++ C + GTP+ N P++F TP DN +++
Sbjct: 656 RDRIYNENNIDS-------LFAKARQKNCPKGSSGTPKDNNVAPLDF-KTPNHFDNEYFK 811
Query: 321 DLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEV 380
+L++K K LL +D L T V +++Q +F F M K+ ++ LTG++G++
Sbjct: 812 NLINK-KGLLRSDQELFNGGSTDSLVRTYSNNQRVFEADFVTAMIKMGNIKPLTGSNGQI 988
Query: 381 RKICRATN 388
RK CR N
Sbjct: 989 RKQCRRPN 1012
>TC204493 UP|O22443 (O22443) Seed coat peroxidase precursor , complete
Length = 1294
Score = 186 bits (471), Expect = 2e-47
Identities = 110/304 (36%), Positives = 165/304 (54%), Gaps = 3/304 (0%)
Frame = +1
Query: 88 FYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNGDNVE 147
FY ++CPN I+ + T+P+ A+++RL FHDCFV GCD S+LL++T ++ E
Sbjct: 112 FYRETCPNLFPIVFGVIFDASFTDPRIGASLMRLHFHDCFVQGCDGSVLLNNTDTIES-E 288
Query: 148 KSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPLGGRR 207
+ + N ++G D+V+DIK+ +E CP VSCAD + + A L G P GRR
Sbjct: 289 QDALPNINSIRGLDVVNDIKTAVENSCPDTVSCADILAIAAEIASVLGGGPGWPVPLGRR 468
Query: 208 DSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIFMDRV 267
DSL + T+ + N LP P ++ ++ F +G + ++V L G H+ G A C F++R+
Sbjct: 469 DSLTANRTLANQN-LPAPFFNLTQLKASFAVQGLNTLDLVTLSGGHTFGRARCSTFINRL 645
Query: 268 YNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNLFYRDLVDKG 326
YNF NT PDP L +L LR C T + N D +TP DN +Y +L+
Sbjct: 646 YNFSNTGNPDPTLNTTYLEVLRARCPQNAT---GDNLTNLDLSTPDQFDNRYYSNLLQL- 813
Query: 327 KSLLLTDAHLVTDP--RTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRKIC 384
LL +D L + P T P V + +Q F F M K+ ++ VLTG++GE+R C
Sbjct: 814 NGLLQSDQELFSTPGADTIPIVNSFSSNQNTFFSNFRVSMIKMGNIGVLTGDEGEIRLQC 993
Query: 385 RATN 388
N
Sbjct: 994 NFVN 1005
>TC206281 UP|Q9ZRG5 (Q9ZRG5) Peroxidase (Fragment) , complete
Length = 1129
Score = 186 bits (471), Expect = 2e-47
Identities = 108/306 (35%), Positives = 167/306 (54%)
Frame = +3
Query: 83 NLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPN 142
+L +YS +CP+ E I+A A + + A +LR+ FHDCFV GCDAS+LL+S
Sbjct: 102 SLSLNYYSKTCPDVECIVAKAVKDATARDKTVPAALLRMHFHDCFVRGCDASVLLNS--K 275
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKP 202
G N + + L ++D K LE CPG+VSCAD + + +A+ L+G P
Sbjct: 276 GSNKAEKDGPPNVSLHAFYVIDAAKKALEASCPGVVSCADILALAARDAVFLSGGPTWDV 455
Query: 203 LGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
GR+D S A+ + LP P ++ ++ + F ++G S E++V L G H++G +HC
Sbjct: 456 PKGRKDGRTSKAS--ETRQLPAPTFNLSQLRQSFSQRGLSGEDLVALSGGHTLGFSHCSS 629
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNLFYRDL 322
F +R++NF T+ DP+L P F +L IC P + +N + D + T DN +YR L
Sbjct: 630 FKNRIHNFNATHDVDPSLNPSFATKLISIC--PLKNQAKNAGTSMDPSTTTFDNTYYR-L 800
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRK 382
+ + K L +D L+ +P T V + A + F+ FA+ M K++S+N EVRK
Sbjct: 801 ILQQKGLFSSDQVLLDNPDTKNLVAKFATSKKAFYDAFAKSMIKMSSIN----GGQEVRK 968
Query: 383 ICRATN 388
CR N
Sbjct: 969 DCRVIN 986
>TC206280 UP|Q9ZRG6 (Q9ZRG6) Peroxidase (Fragment) , complete
Length = 1174
Score = 183 bits (465), Expect = 1e-46
Identities = 108/311 (34%), Positives = 170/311 (53%)
Frame = +3
Query: 78 NTINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILL 137
+T +L +Y+ +CPN E I+A A + + A ILR+ FHDCFV GCDAS+LL
Sbjct: 108 STTGKSLSLNYYAKTCPNVEFIVAKAVKDATARDKTVPAAILRMHFHDCFVRGCDASVLL 287
Query: 138 DSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGM 197
+S G+N + + L ++ K LE CPG+VSCAD + + +A+ L+G
Sbjct: 288 NS--KGNNKAEKDGPPNVSLHAFYVIVAAKKALEASCPGVVSCADILALAARDAVFLSGG 461
Query: 198 PRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGA 257
P GR+D S A+ + LP P ++ ++ + F ++G S E++V L G H++G
Sbjct: 462 PTWDVPKGRKDGRTSKAS--ETRQLPAPTFNLSQLRQSFSQRGLSGEDLVALSGGHTLGF 635
Query: 258 AHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVMDNL 317
+HC F +R++NF T+ DP+L P F +L IC P + +N + D + T DN
Sbjct: 636 SHCSSFKNRIHNFNATHDVDPSLNPSFAAKLISIC--PLKNQAKNAGTSMDPSTTTFDNT 809
Query: 318 FYRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGND 377
+YR L+ + K L +D L+ +P T V + A + F++ FA+ M +++S+N
Sbjct: 810 YYR-LILQQKGLFSSDQVLLDNPDTKNLVTKFATSKKAFYEAFAKSMIRMSSIN----GG 974
Query: 378 GEVRKICRATN 388
EVRK CR N
Sbjct: 975 QEVRKDCRMIN 1007
>TC229562 similar to GB|CAA70034.1|1620369|ATP22A peroxidase ATP22a
{Arabidopsis thaliana;} , partial (93%)
Length = 1233
Score = 183 bits (465), Expect = 1e-46
Identities = 114/309 (36%), Positives = 164/309 (52%), Gaps = 4/309 (1%)
Frame = +2
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILL-DSTPN 142
L GFY ++CPN E+++ A A+ + LRL FHDCFV GCDASILL + P
Sbjct: 104 LSSGFYKNTCPNVEQLVRSAVAQKFQQTFVTAPATLRLFFHDCFVRGCDASILLANGRPE 283
Query: 143 GDNVEKSSFFNGILLKGPDLVDDIKSKLEEE--CPGIVSCADTMVFLSFEAMALAGMPRQ 200
D+ ++ S G D V K+ ++ + C VSCAD + + + + LAG P
Sbjct: 284 KDHPDQISLAGD----GFDTVIKAKAAVDRDPKCRNKVSCADILALATRDVVNLAGGPFY 451
Query: 201 KPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHC 260
GRRD S V +LP P ++ D++ +F G S +M+ L GAH+IG +HC
Sbjct: 452 NVELGRRDGRISTIASVQ-RHLPHPEFNLDQLNSMFNFNGLSQTDMIALSGAHTIGFSHC 628
Query: 261 DIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDN-TPTVMDNLFY 319
+ F +R+YNF N+ DP L + +LRQ+C PR +N D TP DN ++
Sbjct: 629 NKFSNRIYNFSPRNRIDPTLNLQYAFQLRQMCPLRVDPRI---AINMDPVTPQKFDNQYF 799
Query: 320 RDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGE 379
++L +GK L +D L TD R+ TV A ++ F K F + +TKL + V TGN GE
Sbjct: 800 KNL-QQGKGLFTSDQVLFTDARSKATVNLFASNEGAFQKAFVDAVTKLGRVGVKTGNQGE 976
Query: 380 VRKICRATN 388
+R C N
Sbjct: 977 IRFDCTRPN 1003
>TC204669 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (81%)
Length = 1413
Score = 181 bits (459), Expect = 5e-46
Identities = 116/314 (36%), Positives = 174/314 (54%), Gaps = 3/314 (0%)
Frame = +2
Query: 78 NTINPN---LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDAS 134
NT+ N R GFYS +CP AE I+ +R++P ILR+ FHDCFV GCDAS
Sbjct: 218 NTVQWNGEGTRVGFYSSTCPRAESIVRSTVESHLRSDPTLAGPILRMHFHDCFVRGCDAS 397
Query: 135 ILLDSTPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMAL 194
+L+ G E+++ N + L+G D++DD K+K+E CPG+VSCAD + + +++ L
Sbjct: 398 VLI----AGAGTERTAGPN-LSLRGFDVIDDAKAKIEALCPGVVSCADILSLAARDSVVL 562
Query: 195 AGMPRQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHS 254
+G + GR+D S+ + + LP PN + + F KG + E++VIL G H+
Sbjct: 563 SGGLSWQVPTGRKDGRVSIGS--EALTLPGPNDTVATQKDKFSNKGLNTEDLVILAGGHT 736
Query: 255 IGAAHCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDNTPTVM 314
IG + C F DR+Y N N DP++ P FL LRQIC P T + ++ +
Sbjct: 737 IGTSACRSFADRIY---NPNGTDPSIDPSFLPFLRQIC--PQTQPTKRVALD-TGSQFKF 898
Query: 315 DNLFYRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLT 374
D ++ LV +G+ +L +D L TD T V Q F +F + M K++++ V T
Sbjct: 899 DTSYFAHLV-RGRGILRSDQVLWTDASTRGFV-QKYLATGPFKVQFGKSMIKMSNIGVKT 1072
Query: 375 GNDGEVRKICRATN 388
G+ GE+RKIC A N
Sbjct: 1073GSQGEIRKICSAIN 1114
>TC219609 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (79%)
Length = 1036
Score = 181 bits (458), Expect = 7e-46
Identities = 114/307 (37%), Positives = 169/307 (54%), Gaps = 5/307 (1%)
Frame = +2
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L FY SCP A I VR + A++LRL FHDCFV GCDAS+LLD T N
Sbjct: 152 LSSKFYDKSCPKALTTIRKEVERAVRNESRMGASLLRLHFHDCFVQGCDASVLLDDTANF 331
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EK+SF N L+G +++D+IKSKLE C G+VSCAD + + +A+ G + +
Sbjct: 332 TG-EKNSFPNANSLRGFEVIDNIKSKLEGMCKGVVSCADILAVAARDAVVALGGQKWEVQ 508
Query: 204 GGRRDSLYSLATVVDDN-NLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDI 262
GRRDS + A++ + N +LP P ++ F +K F+ +E+V L G H+IG C
Sbjct: 509 VGRRDS--TTASLDEANSDLPAPFLDLSGLITAFAKKNFTTQELVTLSGGHTIGLVRCRF 682
Query: 263 FMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFDN-TPTVMDNLFYRD 321
F R+YN N + P F +++ +C G + FD+ TP DN FY++
Sbjct: 683 FRARIYNESN-------IDPTFAQQMQALCPFEGGD---DNLSPFDSTTPFKFDNAFYKN 832
Query: 322 LVDKGKSLLLTDAHLVTDPRTAPT---VGQMADDQALFHKRFAEVMTKLTSLNVLTGNDG 378
LV K ++ +D L T+ + PT V + + + F K FA+ M K++ L LTG++G
Sbjct: 833 LVQL-KGVVHSDQQLFTNNGSGPTNDQVNRYSRNMGNFKKDFADAMFKMSMLTPLTGSNG 1009
Query: 379 EVRKICR 385
++R+ CR
Sbjct: 1010QIRQNCR 1030
>TC226717 homologue to UP|Q9ZTW8 (Q9ZTW8) Peroxidase, partial (85%)
Length = 1343
Score = 181 bits (458), Expect = 7e-46
Identities = 109/306 (35%), Positives = 154/306 (49%), Gaps = 1/306 (0%)
Frame = +3
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ GFY+ SCP AE+II E + P A ++RL FHDCFV GCD S+L+DSTP G
Sbjct: 162 LQLGFYAKSCPKAEQIILKYVVEHIHNAPSLAAALIRLHFHDCFVNGCDGSVLVDSTP-G 338
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
+ EK + N + L+G ++ IK +E ECPG+VSCAD + + +++ G P
Sbjct: 339 NQAEKDAIPN-LTLRGFGFIEAIKRLVEAECPGVVSCADILALTARDSIHATGGPYWNVP 515
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GRRD S A +LP P + + LF G ++V+L+GAH+IG AHC
Sbjct: 516 TGRRDGFISRA-ADPLRSLPAPFHNLTTQLTLFGNVGLDANDLVLLVGAHTIGIAHCSSI 692
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQI-CSNPGTPRYRNEPVNFDNTPTVMDNLFYRDL 322
R+YNF DP + + L+ C N N + D +L Y
Sbjct: 693 STRLYNFTGKGDTDPTIDNGYAKNLKTFKCKNIND----NSLIEMDPGSRDTFDLGYYKQ 860
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGNDGEVRK 382
V K + L +DA L+T P T + F FA+ M K+ +NV G++GE+RK
Sbjct: 861 VVKRRGLFQSDAELLTSPITRSIIASQLQSTQGFFAEFAKSMEKMGRINVKLGSEGEIRK 1040
Query: 383 ICRATN 388
C N
Sbjct: 1041HCARVN 1058
>TC206016 similar to UP|Q6T1C8 (Q6T1C8) Peroxidase precursor, partial (95%)
Length = 1214
Score = 181 bits (458), Expect = 7e-46
Identities = 112/311 (36%), Positives = 162/311 (52%), Gaps = 1/311 (0%)
Frame = +3
Query: 79 TINPNLREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLD 138
T + NL + FYS +CPN + V P+ A+I+RL FHDCFV GCD SILLD
Sbjct: 102 TSSANLSKNFYSKTCPNVFNTVKSVVKSAVVREPRIGASIVRLFFHDCFVQGCDGSILLD 281
Query: 139 STPNGDNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMP 198
TP EK++ N ++G +++D IKS++E+ CPG+VSCAD + S +++ L G P
Sbjct: 282 DTPTFQG-EKTAAANNNSVRGFEVIDAIKSEVEKICPGVVSCADILDIASRDSVVLLGGP 458
Query: 199 RQKPLGGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAA 258
K GRRDS + T + +P P + ++ FQ +G S +MV L GAH+ G A
Sbjct: 459 FWKVRLGRRDSRTANFTAANTGVIPPPTSNLTNLITRFQDQGLSARDMVALSGAHTFGKA 638
Query: 259 HCDIFMDRVYNFKNTNKPDPALRPPFLNELRQICSNPGTPRYRNEPVNFD-NTPTVMDNL 317
C F DR+YN N ++ R R+ GT N N D TP DN
Sbjct: 639 RCTSFRDRIYNQTNIDRTFALARQ------RRCPRTNGTG--DNNLANLDFRTPNHFDNN 794
Query: 318 FYRDLVDKGKSLLLTDAHLVTDPRTAPTVGQMADDQALFHKRFAEVMTKLTSLNVLTGND 377
++++L+ K + LL +D L T V + + F F + M ++ + LTG+
Sbjct: 795 YFKNLLIK-RGLLNSDQVLFNGGSTDSLVRTYSQNNKAFDTDFVKAMIRMGDIKPLTGSQ 971
Query: 378 GEVRKICRATN 388
GE+RK CR N
Sbjct: 972 GEIRKNCRRVN 1004
>TC215016 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
complete
Length = 1453
Score = 180 bits (456), Expect = 1e-45
Identities = 110/307 (35%), Positives = 165/307 (52%), Gaps = 2/307 (0%)
Frame = +3
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ GFY+ SCP AE+II E + P A ++R+ FHDCFV GCDAS+LL+ST N
Sbjct: 219 LQLGFYAKSCPKAEQIILKFVHEHIHNAPSLAAALIRMHFHDCFVRGCDASVLLNSTTN- 395
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EK++ N + ++G D +D IKS +E ECPG+VSCAD + + + + G P K
Sbjct: 396 -QAEKNAPPN-LTVRGFDFIDRIKSLVEAECPGVVSCADILTLSARDTIVATGGPFWKVP 569
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GRRD + S T D N+P P+ + + LF +G +++V+L GAH+IG AHC
Sbjct: 570 TGRRDGVISNLTEARD-NIPAPSSNFTTLQTLFANQGLDLKDLVLLSGAHTIGIAHCSSL 746
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQI-CSNPGTPRYRNEPVNFDNTPTVMDNLFYRDL 322
+R++NF DP+L + L+ C++ + + D +L Y
Sbjct: 747 SNRLFNFTGKGDQDPSLDSEYAANLKAFKCTD--LNKLNTTKIEMDPGSRKTFDLSYYSH 920
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQA-LFHKRFAEVMTKLTSLNVLTGNDGEVR 381
V K + L +DA L+T+ T + ++ + F FA M K+ +NV TG +GE+R
Sbjct: 921 VIKRRGLFESDAALLTNSVTKAQIIELLEGSVENFFAEFATSMEKMGRINVKTGTEGEIR 1100
Query: 382 KICRATN 388
K C N
Sbjct: 1101KHCAFVN 1121
>TC215014 homologue to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
complete
Length = 1341
Score = 180 bits (456), Expect = 1e-45
Identities = 110/307 (35%), Positives = 164/307 (52%), Gaps = 2/307 (0%)
Frame = +1
Query: 84 LREGFYSDSCPNAEKIIADAFAEIVRTNPKAMANILRLQFHDCFVVGCDASILLDSTPNG 143
L+ GFY+ SCP AEKII E + P A ++R+ FHDCFV GCD S+LL+ST N
Sbjct: 163 LQLGFYAQSCPKAEKIILKFVHEHIHNAPSLAAALIRMHFHDCFVRGCDGSVLLNSTTN- 339
Query: 144 DNVEKSSFFNGILLKGPDLVDDIKSKLEEECPGIVSCADTMVFLSFEAMALAGMPRQKPL 203
EK++ N + ++G D +D IKS +E ECPG+VSCAD + + + + G P K
Sbjct: 340 -QAEKNAPPN-LTVRGFDFIDRIKSLVEAECPGVVSCADILTLAARDTIVATGGPFWKVP 513
Query: 204 GGRRDSLYSLATVVDDNNLPMPNWSADKMVELFQRKGFSPEEMVILLGAHSIGAAHCDIF 263
GRRD + S T NN+P P+ + + LF +G +++V+L GAH+IG AHC
Sbjct: 514 TGRRDGVVSNLTEA-RNNIPAPSSNFTTLQTLFANQGLDLKDLVLLSGAHTIGIAHCSSL 690
Query: 264 MDRVYNFKNTNKPDPALRPPFLNELRQI-CSNPGTPRYRNEPVNFDNTPTVMDNLFYRDL 322
+R++NF DP+L + L+ C++ + + D +L Y
Sbjct: 691 SNRLFNFTGKGDQDPSLDSEYAANLKAFKCTD--LNKLNTTKIEMDPGSRKTFDLSYYSH 864
Query: 323 VDKGKSLLLTDAHLVTDPRTAPTVGQMADDQA-LFHKRFAEVMTKLTSLNVLTGNDGEVR 381
V K + L +DA L+T+ T + Q+ + F FA + K+ +NV TG +GE+R
Sbjct: 865 VIKRRGLFESDAALLTNSVTKAQIIQLLEGSVENFFAEFATSIEKMGRINVKTGTEGEIR 1044
Query: 382 KICRATN 388
K C N
Sbjct: 1045KHCAFIN 1065
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,800,567
Number of Sequences: 63676
Number of extensions: 242095
Number of successful extensions: 1462
Number of sequences better than 10.0: 191
Number of HSP's better than 10.0 without gapping: 1241
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1281
length of query: 388
length of database: 12,639,632
effective HSP length: 99
effective length of query: 289
effective length of database: 6,335,708
effective search space: 1831019612
effective search space used: 1831019612
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0076a.9


