

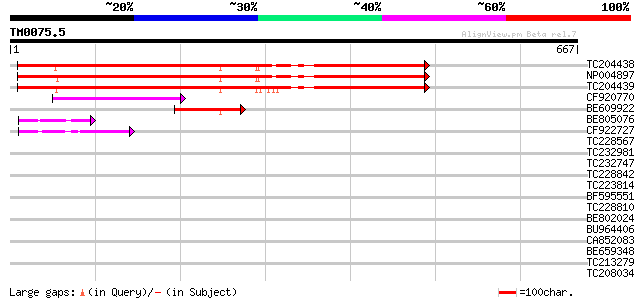
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0075.5
(667 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 469 e-132
NP004897 gag-protease polyprotein 459 e-129
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 451 e-127
CF920770 115 8e-26
BE609922 similar to GP|29423273|g gag-pol polyprotein {Glycine m... 63 4e-10
BE805076 weakly similar to GP|9294121|dbj copia-like retrotransp... 47 2e-05
CF922727 44 2e-04
TC228567 40 0.004
TC232981 UP|Q9LLR0 (Q9LLR0) Carboxyl transferase alpha subunit ... 36 0.048
TC232747 35 0.11
TC228842 34 0.18
TC223814 33 0.41
BF595551 similar to GP|16974544|gb| At2g03500/T4M8.7 {Arabidopsi... 32 0.69
TC228810 weakly similar to UP|Q9LIQ9 (Q9LIQ9) Arabidopsis thalia... 32 0.90
BE802024 31 2.0
BU964406 30 2.6
CA852083 30 2.6
BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor p... 30 2.6
TC213279 similar to UP|TRSF_DROHY (Q23949) Female-specific trans... 30 3.4
TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 30 3.4
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 469 bits (1207), Expect = e-132
Identities = 258/538 (47%), Positives = 341/538 (62%), Gaps = 54/538 (10%)
Frame = +1
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIAST-----TELKPEDKWTKK 64
PPILDGTNY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 37 PPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKE 216
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+AWEILKT HEGTSKV+MS+LQLL T
Sbjct: 217 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLAT 396
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE M++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 397 KFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEE 576
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 577 AQDICNMRVDELIGSLQTFELGLSDRTEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVV 756
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI---------HVK 291
LL NK L + R +V + D K +E+Q K + +K I H+K
Sbjct: 757 LLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKKSDEKPSHSKGIQCHGCEGYGHIK 936
Query: 292 -----------------------------------ALIGKCYSDAESSDGDEEELVETYK 316
AL G+ S +SSD D E T+
Sbjct: 937 AECPTHLKKQRKGLSVCRSDDTESEQESDSDRDVNALTGRFESAEDSSDTDSE---ITFD 1107
Query: 317 LLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAK 376
L + E C+ EK ++ ++ QL+ K I+ L E E + ++
Sbjct: 1108ELAISYRELCIKSEK-------ILQQEAQLK----------KVIANLEAEKEAHEEEISE 1236
Query: 377 LQEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE- 435
L+ EV LNSKLE M KSI+M+NK +++L+E+L++GK VG+ G+GF++KSA ++ +E
Sbjct: 1237LKGEVGFLNSKLENMTKSIKMLNKGSDMLDEVLQLGKNVGNQRGLGFNHKSAGRTTMTEF 1416
Query: 436 KQTKQPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
K M H RH Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 1417VPAKNSTGATMSQHRSRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 1590
>NP004897 gag-protease polyprotein
Length = 1923
Score = 459 bits (1181), Expect = e-129
Identities = 254/538 (47%), Positives = 337/538 (62%), Gaps = 54/538 (10%)
Frame = +1
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE-----LKPEDKWTKK 64
PPILDGTNY+YWKARM+ FLKS+DS WKA++K W+HP + T LKPE+ WTK+
Sbjct: 37 PPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKDWEHPKMLDTEGKPTDGLKPEEDWTKE 216
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+AWEILKT HEGTSKV+MS+LQLL T
Sbjct: 217 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLAT 396
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE M++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 397 KFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEE 576
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 577 AQDICNLRVDELIGSLQTFELGLSDRTEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVV 756
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI---------HVK 291
LL NK L + R +V + D K +E+Q + + +K H+K
Sbjct: 757 LLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKRSDEKPSHSKGFQCHGCEGYGHIK 936
Query: 292 -----------------------------------ALIGKCYSDAESSDGDEEELVETYK 316
AL G+ S +SSD D E T+
Sbjct: 937 AECPTHLKKQRKGLSVCRSDDTESEQESDSDRDVNALTGRFESAEDSSDTDSE---ITFD 1107
Query: 317 LLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAK 376
L + E C+ EK ++ ++ QL+ K I+ L E E + ++
Sbjct: 1108ELATSYRELCIKSEK-------ILQQEAQLK----------KVIANLEAEKEAHEEEISE 1236
Query: 377 LQEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE- 435
L+ EV LNSKLE M KSI+M+NK +++L+E+L++GK VG+ G+GF++KSA + +E
Sbjct: 1237LKGEVGFLNSKLENMTKSIKMLNKGSDMLDEVLQLGKNVGNQRGLGFNHKSAGRITMTEF 1416
Query: 436 KQTKQPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
K M H RH Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 1417VPAKISTGATMSQHRSRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 1590
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 451 bits (1160), Expect = e-127
Identities = 252/534 (47%), Positives = 338/534 (63%), Gaps = 50/534 (9%)
Frame = +1
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTT-----ELKPEDKWTKK 64
PPILDG+NY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 37 PPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKE 216
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+AWEILK HEGTSKV+MS+LQLL T
Sbjct: 217 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLAT 396
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE +++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 397 KFENLKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEE 576
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 577 AQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVV 756
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI------------ 288
LL NK L + + +V + D K +++Q + + +K I
Sbjct: 757 LLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGIQCHGCEGYGHII 936
Query: 289 -----HVKAL---IGKCYSDAES---SDGDEE--------ELVE---------TYKLLLA 320
H+K + C SD ES SD D + E E T+ L A
Sbjct: 937 AECPTHLKKHRKGLSVCQSDTESEQESDSDRDVNALTGIFETAEDSSDTDSEITFDELAA 1116
Query: 321 KWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEE 380
+ + C+ EK ++ ++ QL+ K I+ L E E + ++L+ E
Sbjct: 1117SYRKLCIKSEK-------ILQQEAQLK----------KVIADLEAEKEAHKEEISELKGE 1245
Query: 381 VTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE-KQTK 439
V LNSKLE M KSI+M+NK ++ L+E+L +GK G+ G+GF+ KSA ++ +E K
Sbjct: 1246VGFLNSKLENMTKSIKMLNKGSDTLDEVLLLGKNAGNQRGLGFNPKSAGRTTMTEFVPAK 1425
Query: 440 QPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
M H RH Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 1426NRTGATMSQHRSRHHGMQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 1587
>CF920770
Length = 581
Score = 115 bits (287), Expect = 8e-26
Identities = 57/157 (36%), Positives = 90/157 (57%)
Frame = -2
Query: 51 STTELKPEDKWTKKEDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEG 110
S T KP D+W++++ N KA N+I + + + + ++ C AKE W+ L+ HEG
Sbjct: 484 SITIEKPRDRWSEEDRKRVQYNLKAKNIITSALGMDEYFRVSNCKSAKEMWDTLRLTHEG 305
Query: 111 TSKVRMSKLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRS 170
T+ V+ S++ LT ++E +MN +E+I R + N ALG+ E L K+LR
Sbjct: 304 TTDVKRSRINALTHEYELFRMNTNENIQSMQKRFTHIVNHLAALGKEFQNEDLINKVLRC 125
Query: 171 LPKRFDMKVTAIEEAQDISNIKVDELIGSLQTFEMSL 207
L + + KVTAI E++D+SN+ + L G LQ EM L
Sbjct: 124 LSREWQPKVTAISESRDLSNMSLATLFGKLQEHEMEL 14
>BE609922 similar to GP|29423273|g gag-pol polyprotein {Glycine max}, partial
(7%)
Length = 340
Score = 63.2 bits (152), Expect = 4e-10
Identities = 34/88 (38%), Positives = 54/88 (60%), Gaps = 4/88 (4%)
Frame = +2
Query: 194 DELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLL----NKA 249
DEL+GSLQTFE+ L+ R+EKK+K++ F+SN E +E + + DTD + A LL +K
Sbjct: 2 DELMGSLQTFELGLSDRAEKKSKNLAFMSNDEGEEGEYDLDTDEGLTNAGVLLGKQFSKV 181
Query: 250 LKSLGRMSNTNVLDNVSDNVKNTEFQLK 277
+ + R +V D K++++Q K
Sbjct: 182 MNRIDRRQKPHVQSRRGDVRKSSKYQRK 265
>BE805076 weakly similar to GP|9294121|dbj copia-like retrotransposable
element {Arabidopsis thaliana}, partial (2%)
Length = 388
Score = 47.4 bits (111), Expect = 2e-05
Identities = 28/91 (30%), Positives = 40/91 (43%)
Frame = +1
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEAL 70
PI +G NYD+W+ +M +L S D W + +G+ P S E + K +
Sbjct: 133 PIFNGENYDFWRVKMETYLSSQD--LWDIVEEGFTIPADTSALNASQEKELKKNKQK--- 297
Query: 71 GNSKALNVIFNGVDKNMFRLINTCTVAKEAW 101
NSK L + +F I AKEAW
Sbjct: 298 -NSKTLFTLQQAETDPIFPRIMGAKTAKEAW 387
>CF922727
Length = 791
Score = 43.9 bits (102), Expect = 2e-04
Identities = 39/138 (28%), Positives = 62/138 (44%), Gaps = 1/138 (0%)
Frame = -2
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEAL 70
PIL G NY WK R+++ L MD ++ + PVI T+E D + K E
Sbjct: 430 PILKGDNYKVWKERVLLHLGWMDI---DYAIRKDEPPVITETSEPDVVDLYEKWE----- 275
Query: 71 GNSKALNVIFNGVDKNMFRLI-NTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETM 129
S L+V+F + N+ I + + ++LK E + S L QF ++
Sbjct: 274 -RSNRLSVMF--IKTNISTSIWGSVDQHDKVRDLLKAIDERFTTSEKSLASTLIMQFSSI 104
Query: 130 KMNEDESIYEFHMRIRDL 147
K+ + E MR+RD+
Sbjct: 103 KLTGTRGVREHIMRLRDI 50
>TC228567
Length = 1531
Score = 39.7 bits (91), Expect = 0.004
Identities = 72/358 (20%), Positives = 141/358 (39%), Gaps = 14/358 (3%)
Frame = +2
Query: 67 DEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQF 126
D A+GN++ ++K L KEAW ++ AH +V+ +L + +
Sbjct: 17 DIAIGNAEKTIAASKQIEKAAEDLTAEFIATKEAWNSMRPAH---*EVKQQRL*AVDQES 187
Query: 127 ETMKMNEDESIYEFHMRIRDLANSTFALGEPM-SEEKLARKILRSLPKRFDMKVTAIEEA 185
+K+ E E + E R+ + ST L + S L + L + KV EE
Sbjct: 188 NNLKL-ELEQV*EELQRLNEQVLSTRVLKSKLESASSLLHGLKAELTAYMESKVN--EEF 358
Query: 186 QDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDED------QREKDTDANI 239
+V+EL +++ +N + S++ S EE++ Q E+ A +
Sbjct: 359 YKEQKEEVEELKLNIEKATSDVN---RLRVASVSLKSKLEEEKSVLASLKQSEEKASAAV 529
Query: 240 AEAVSLLNKALKSLG--RMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAIHVKALIGKC 297
+ L K+ ++ +M + +++ K + K E D K++ A
Sbjct: 530 VNLQAELEKSRSAIAFIQMKENEAREMMTELPKKLQ---KASQEADEAKSLAQAAQAELI 700
Query: 298 YSDAESSDGDEEELVETYKLLLAKWE-ESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEE 356
+ E + LL A+ E E+ E + ++ + EK + N + ++
Sbjct: 701 EAQEEVEQAKAKSSTLESSLLAAQKEIEAAKVAEMLARDAITAL-EKSESAKGNKNDKDS 877
Query: 357 VKTISKLREENEKLQITNAKLQEEVTLLNSKLEGMKKSIRMMN----KSTNVLEEILE 410
++ EE +L K +E+ N+++E I++ +S LEE+ E
Sbjct: 878 SSMVTLTLEEYHELSRRAYKAEEQA---NARIEAATSQIQIARESELRSLEKLEELNE 1042
Score = 38.1 bits (87), Expect = 0.013
Identities = 62/274 (22%), Positives = 114/274 (40%), Gaps = 3/274 (1%)
Frame = +2
Query: 117 SKLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFD 176
SKL+ + ++K +E+++ +L S A+ +E AR+++ LPK+
Sbjct: 458 SKLEEEKSVLASLKQSEEKASAAVVNLQAELEKSRSAIAFIQMKENEAREMMTELPKKLQ 637
Query: 177 MKVTAIEEAQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTD 236
+EA+ ++ ELI + + E + KAKS T S+ + + E
Sbjct: 638 KASQEADEAKSLAQAAQAELIEAQEEVE-------QAKAKSSTLESSLLAAQKEIEA--- 787
Query: 237 ANIAEAVSLLNKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAIHVKALIGK 296
A +AE L A+ +L + + N D+ L++ HE L +
Sbjct: 788 AKVAE--MLARDAITALEKSESAKGNKNDKDSSSMVTLTLEEYHE-----------LSRR 928
Query: 297 CYSDAESSDGDEEELVETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQ--SNNSSLQ 354
Y E ++ E T ++ +A+ E EK+ + ++L ++ L+ + NS
Sbjct: 929 AYKAEEQANARIE--AATSQIQIAR-ESELRSLEKLEELNEELSVRRESLKIATGNSEKA 1099
Query: 355 EEVKTISKLREENE-KLQITNAKLQEEVTLLNSK 387
E KL E+E + K QE+ T LN +
Sbjct: 1100NE----GKLAVEHELRTWRAEQKQQEKATELNEQ 1189
>TC232981 UP|Q9LLR0 (Q9LLR0) Carboxyl transferase alpha subunit , complete
Length = 2799
Score = 36.2 bits (82), Expect = 0.048
Identities = 64/296 (21%), Positives = 124/296 (41%), Gaps = 24/296 (8%)
Frame = +3
Query: 118 KLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDM 177
K + T E KMN +E + ++ R + F G P+ ++ A K+ D+
Sbjct: 1452 KKAIKETMDELTKMNTEELLKHRMLKFRKIGG--FQEGIPIDPKRKANM------KKRDL 1607
Query: 178 KVTAIEEAQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDA 237
+ I +A+ ++V++L Q E K++ + + +E Q ++ D
Sbjct: 1608 SIAKIPDAE--LEVEVEKL--KQQVLEA-------KESSPVPPKLDLDEMLKQLAREVDL 1754
Query: 238 NIAEAVS---LLNKALKSLGRMSNTNVLDNVSDNVKNTEFQ-LKDKHENDTTKAIHVKAL 293
+EAV L + LK +S N + + D + + + L+ + E A + L
Sbjct: 1755 EYSEAVKATGLTDSLLKLREEVSKANADNQIVDPLLEGKIEKLRVEFEQQLRAAPNYGRL 1934
Query: 294 IGKCYSDAES------SDGDEEELVETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQ 347
K +E SDG + T+K L K ++ + K+R+ + L AE K +
Sbjct: 1935 QNKLNYLSELCKVKLLSDGKKNNEAVTFKQELKKKIDNALSDPKIRETFEALKAEIKGVG 2114
Query: 348 SNNSS------------LQEEVKTISKLRE--ENEKLQITNAKLQEEVTLLNSKLE 389
++++S +EVK + +++E EN+ + N+ + +L KLE
Sbjct: 2115 ASSASDLDDELKKKIIEFIKEVKEVKEVKEVIENQIESLVNSSDDIKSKILQLKLE 2282
>TC232747
Length = 681
Score = 35.0 bits (79), Expect = 0.11
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 4/78 (5%)
Frame = +3
Query: 316 KLLLAKWEESCMYGE----KMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQ 371
+LL+AK +E + ++R E L+ ++K+ + N +S LREENE LQ
Sbjct: 15 ELLIAKTKEVAAKSDAELGQVRSERNKLVEKEKEREGN----------VSVLREENEALQ 164
Query: 372 ITNAKLQEEVTLLNSKLE 389
AK ++E LN+K+E
Sbjct: 165 GMLAKARKESKDLNAKVE 218
Score = 28.9 bits (63), Expect = 7.6
Identities = 34/152 (22%), Positives = 69/152 (45%), Gaps = 22/152 (14%)
Frame = +3
Query: 288 IHVKALIGKCYSDAESSDGD-------EEELVETYK-----LLLAKWEESCMYG--EKMR 333
+ ++ LI K A SD + +LVE K + + + E + G K R
Sbjct: 6 VKLELLIAKTKEVAAKSDAELGQVRSERNKLVEKEKEREGNVSVLREENEALQGMLAKAR 185
Query: 334 KEVKDLIAEKKQLQSNNSSLQEEVKTIS-------KLREENEKLQITNAKLQEEVTLLNS 386
KE KDL A+ + SN++ +KT + K R +E + + EE+
Sbjct: 186 KESKDLNAKVEVWCSNSNKALSLLKTTAAALVCQHKERGGDEVVAAADENPVEEIQPYAQ 365
Query: 387 KLEGMKKSIRMMNKST-NVLEEILEVGKTVGD 417
+L+ +KK+ + ++ ++ ++++ + K+V +
Sbjct: 366 ELDAIKKAFKTKDEMVDDMKQQLVSLNKSVAE 461
>TC228842
Length = 1023
Score = 34.3 bits (77), Expect = 0.18
Identities = 26/90 (28%), Positives = 47/90 (51%), Gaps = 9/90 (10%)
Frame = +2
Query: 330 EKMRK--------EVKDLIAEKKQLQSNNSS-LQEEVKTISKLREENEKLQITNAKLQEE 380
EKMR+ E+ ++ +K L+ + + L + V+ +S+LREE +KL+ + LQE+
Sbjct: 335 EKMRRDRLNDRFMELGSILDPRKPLKMDKAVILSDAVRVVSQLREEAQKLRESTENLQEK 514
Query: 381 VTLLNSKLEGMKKSIRMMNKSTNVLEEILE 410
+ L + K +R + V +E LE
Sbjct: 515 INALKDE----KNELRDEKQRLKVEKENLE 592
>TC223814
Length = 607
Score = 33.1 bits (74), Expect = 0.41
Identities = 47/205 (22%), Positives = 73/205 (34%), Gaps = 1/205 (0%)
Frame = +1
Query: 15 GTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEALGNSK 74
G NYD WK R++ + W + G P+ E P WTK + SK
Sbjct: 7 GQNYDNWKQRIIELFDACHIDMWDVVENGNYIPIDEKGGE-NPRSAWTKDHQTKYFLYSK 183
Query: 75 ALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKMNED 134
A N R N C + I M+ L+ L+ + N D
Sbjct: 184 ARNT----------RPSNNCLDNSKVQNI------------MNNLRSLSKT*D----NHD 285
Query: 135 ESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDISNIKVD 194
+ I KIL+SL + V A+ +++D+ ++ V+
Sbjct: 286 DHI---------------------------TKILQSLLIQ*RP*VIALCDSKDLKSLPVE 384
Query: 195 ELIGSLQTFEMSL-NGRSEKKAKSI 218
E G+LQ E+ L ++K K I
Sbjct: 385 EFDGTLQVHELELMEDEGQRKGKFI 459
>BF595551 similar to GP|16974544|gb| At2g03500/T4M8.7 {Arabidopsis thaliana},
partial (10%)
Length = 418
Score = 32.3 bits (72), Expect = 0.69
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 4/78 (5%)
Frame = +3
Query: 344 KQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSKLEGMK----KSIRMMN 399
+ L +NN+ +S+L +E K+ +L + LLN + +K K R+
Sbjct: 135 RHLSANNNHPATLRDFLSRLEDELRKIHAFKRELPLSMLLLNDAISVLKVESQKCCRVAR 314
Query: 400 KSTNVLEEILEVGKTVGD 417
S VLEE + + K +GD
Sbjct: 315 DSPPVLEEFIPLKKELGD 368
>TC228810 weakly similar to UP|Q9LIQ9 (Q9LIQ9) Arabidopsis thaliana genomic
DNA, chromosome 3, BAC clone:F14O13, partial (30%)
Length = 1277
Score = 32.0 bits (71), Expect = 0.90
Identities = 22/65 (33%), Positives = 32/65 (48%), Gaps = 3/65 (4%)
Frame = +3
Query: 343 KKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSKL---EGMKKSIRMMN 399
+KQL++ + K +S L ++ LQ T LQEE +L SKL G+ KSI
Sbjct: 51 EKQLENAKEEISSYRKKMSSLDKDRHDLQSTIEALQEEKKMLLSKLRKASGIGKSIENQT 230
Query: 400 KSTNV 404
+V
Sbjct: 231 SKRDV 245
>BE802024
Length = 267
Score = 30.8 bits (68), Expect = 2.0
Identities = 22/87 (25%), Positives = 45/87 (51%)
Frame = +3
Query: 333 RKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSKLEGMK 392
+ V+++ EK+QL+ + L E+ LQ+ N ++EVTL S +
Sbjct: 18 KDHVREMKREKEQLKRDKRHLLEQ-------------LQVKN---EQEVTLRKSVEKLEA 149
Query: 393 KSIRMMNKSTNVLEEILEVGKTVGDME 419
K+ + ++ N+ ++++ KTVG++E
Sbjct: 150 KASKEESEKMNLTTTVVQLKKTVGELE 230
>BU964406
Length = 447
Score = 30.4 bits (67), Expect = 2.6
Identities = 24/87 (27%), Positives = 38/87 (43%)
Frame = +3
Query: 427 SANKSASSEKQTKQPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYG 486
S + S SS Q P S+P++ + ++ ++ST CH+C + GH YC
Sbjct: 36 SFSHSPSSSSQAHSPQSNPLIPANC--CLELGKRMQQST--CHNCDQRGHWSYYC----- 188
Query: 487 YPQSHDQPRTNPQVAPTRKEWKPSGLK 513
P+T P +P + PS K
Sbjct: 189 ---PSKSPKTKP-FSPGKDSTVPSNKK 257
>CA852083
Length = 829
Score = 30.4 bits (67), Expect = 2.6
Identities = 35/179 (19%), Positives = 70/179 (38%), Gaps = 15/179 (8%)
Frame = +1
Query: 411 VGKTVGDMEGIGFSYKSANKSASSEKQTKQPMSDPMLHHSVR-HVYPQFRKSKKSTWR-- 467
+G +M+ S K +N + E++ + P + P S V+ K+ + W+
Sbjct: 160 LGTATTNMQERAKSNKRSNAGSDEEEEARDPNAVPTDFTSREAKVWEAKSKATERNWKKR 339
Query: 468 ------CHHCGKLGHIRPYCYKLYGYPQSHDQ-----PRTNPQVAPTRKEWKPSGLKEKE 516
C CG+ GH C G + P + V E S +++
Sbjct: 340 KEEEMICKLCGESGHFTQGCPSTLGANRKSQDFFERIPARDKNVRALFTEKVLSKIEKDV 519
Query: 517 KKEIVKAPRHFLMTNKD-VALLKSQSSGVDVMKEWRKKFVAASSLEGSNAPSTVHASIG 574
+I + +++ KD + L K +G + +E ++ ++S + S +P V +G
Sbjct: 520 GCKIKMDEKFIIVSGKDRLILAKGVDAGHKIREEGDQRGSSSSQMTQSRSPEEVLLVLG 696
>BE659348 weakly similar to PIR|JC7809|JC78 sulfakinin receptor protein
DSK-R1 - fruit fly (Drosophila melanogaster), partial
(6%)
Length = 770
Score = 30.4 bits (67), Expect = 2.6
Identities = 8/21 (38%), Positives = 15/21 (71%)
Frame = -1
Query: 468 CHHCGKLGHIRPYCYKLYGYP 488
C +C ++GH + CY ++G+P
Sbjct: 383 CSYCKRVGHTQDTCYSIHGFP 321
>TC213279 similar to UP|TRSF_DROHY (Q23949) Female-specific transformer
protein, partial (11%)
Length = 481
Score = 30.0 bits (66), Expect = 3.4
Identities = 19/62 (30%), Positives = 33/62 (52%)
Frame = +2
Query: 328 YGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSK 387
YGE+ ++ L E+ +L S N + +EE+K KL E + L+ A E++ L +
Sbjct: 194 YGEESATKIAALQRERDELVSENVARKEEIK---KLTAELDGLRSDGADKSEKIEELQRE 364
Query: 388 LE 389
+E
Sbjct: 365 VE 370
>TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (46%)
Length = 1183
Score = 30.0 bits (66), Expect = 3.4
Identities = 23/103 (22%), Positives = 45/103 (43%), Gaps = 3/103 (2%)
Frame = +3
Query: 323 EESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLRE---ENEKLQITNAKLQE 379
+E + + ++++ +AEK+Q E + +LRE E EK NA+L+
Sbjct: 357 DEIDQFLQAQEEQLRRALAEKRQRHYRTLLRAAEESVLRRLREKEAELEKATRHNAELEA 536
Query: 380 EVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIG 422
T L+ + + + + + L+ L+ +GD E G
Sbjct: 537 RATQLSVEAQLWQARAKAQEATAAALQAQLQQAMMIGDGENGG 665
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,797,531
Number of Sequences: 63676
Number of extensions: 330964
Number of successful extensions: 1684
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 1644
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1670
length of query: 667
length of database: 12,639,632
effective HSP length: 104
effective length of query: 563
effective length of database: 6,017,328
effective search space: 3387755664
effective search space used: 3387755664
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0075.5


