

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
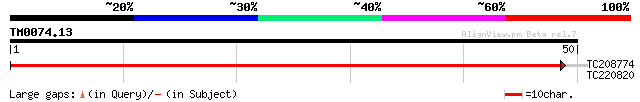
Query= TM0074.13
(50 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208774 weakly similar to UP|Q6GKX4 (Q6GKX4) At5g24170, partial... 91 1e-19
TC220820 similar to UP|O48592 (O48592) GT2 protein, partial (11%) 25 6.6
>TC208774 weakly similar to UP|Q6GKX4 (Q6GKX4) At5g24170, partial (74%)
Length = 887
Score = 90.9 bits (224), Expect = 1e-19
Identities = 43/49 (87%), Positives = 47/49 (95%)
Frame = +1
Query: 1 MEKMSKAFEKVKIMVGMEVEDEEQQAAALDNDNSFAFMDEFNRNCTLST 49
MEKM+KAFEKVKIMVGM+ EDEEQQAAAL+N NSFAFMD+FNRNCTLST
Sbjct: 142 MEKMNKAFEKVKIMVGMDAEDEEQQAAALNNSNSFAFMDDFNRNCTLST 288
>TC220820 similar to UP|O48592 (O48592) GT2 protein, partial (11%)
Length = 646
Score = 25.4 bits (54), Expect = 6.6
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = +3
Query: 22 EEQQAAALDNDNSFAFMDEFNRNCTLS 48
++QQ +LD D F F+D N+ CT S
Sbjct: 366 QQQQVPSLDFDQ-F*FIDSHNKKCTTS 443
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.125 0.327
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,261,463
Number of Sequences: 63676
Number of extensions: 6259
Number of successful extensions: 27
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27
length of query: 50
length of database: 12,639,632
effective HSP length: 26
effective length of query: 24
effective length of database: 10,984,056
effective search space: 263617344
effective search space used: 263617344
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0074.13


