

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.8
(1156 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
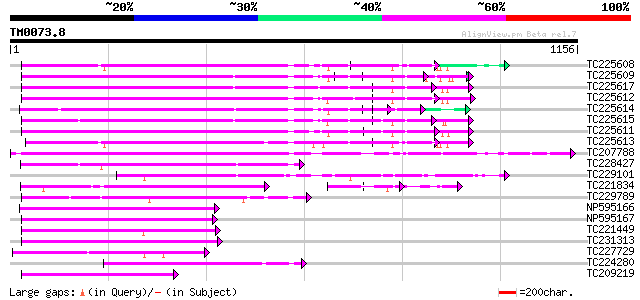
Score E
Sequences producing significant alignments: (bits) Value
TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete 451 e-127
TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete 451 e-127
TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete 448 e-126
TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete 439 e-123
TC225614 UP|Q84ZV7 (Q84ZV7) R 12 protein, partial (88%) 433 e-121
TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete 433 e-121
TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete 427 e-119
TC225613 UP|Q9FVK5 (Q9FVK5) Resistance protein LM6 (Fragment), p... 422 e-118
TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate re... 412 e-115
TC228427 UP|Q8H6S7 (Q8H6S7) Resistance protein KR3, complete 352 4e-97
TC229101 disease resistance-like protein 333 3e-91
TC221834 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candida... 327 1e-89
TC229789 homologue to UP|Q9FVK4 (Q9FVK4) Resistance protein LM17... 288 1e-77
NP595166 R 14 protein [Glycine max] 273 3e-73
NP595167 R 13 protein [Glycine max] 263 3e-70
TC221449 UP|Q9FXZ1 (Q9FXZ1) Resistance protein MG23 (Fragment), ... 255 9e-68
TC231313 UP|Q9FXZ2 (Q9FXZ2) Resistance protein LM12 (Fragment), ... 251 1e-66
TC227729 249 6e-66
TC224280 UP|Q9FVK3 (Q9FVK3) Resistance protein MG63 (Fragment), ... 241 2e-63
TC209219 UP|Q9FVK2 (Q9FVK2) Resistance protein MG13 (Fragment), ... 214 2e-55
>TC225608 UP|Q84ZU5 (Q84ZU5) R 8 protein, complete
Length = 3084
Score = 451 bits (1161), Expect = e-127
Identities = 320/904 (35%), Positives = 475/904 (52%), Gaps = 51/904 (5%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEI----CKAIEASTIYVI 77
YDVFLSF+G+DT + FT +L+ AL + + FIDD+ L GDEI AI+ S I +
Sbjct: 34 YDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S+NYASS CLDELV I+ CK + VIP+FY+VDPS VRHQ SYG+A +HQ R
Sbjct: 214 VLSQNYASSSFCLDELVTILHCK-SQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRF 390
Query: 138 VPGVV-MQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPPFSQGML---- 191
+Q+W+ AL + ADLSGYH S E I I ++I +K FS+ L
Sbjct: 391 KANKEKLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRK----FSRASLHVAD 558
Query: 192 ---GIDKHIAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVN 247
G++ + ++ LL + S + V IIGI GMGG+GK+TLA A+++ + F C + N
Sbjct: 559 YPVGLESEVTEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALHFDESCFLQN 738
Query: 248 AQQKIDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIP 302
+++ +++G+ L+ LSKLLGE DI G + R++R KVLLILDD+
Sbjct: 739 VREESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQ 918
Query: 303 ILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYP 362
+ ++G FG GSR+I+T+R +H+LK E + YEV+ ++ +L+L NAFK +
Sbjct: 919 LKAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLTWNAFKREKI 1098
Query: 363 LEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSY 422
Y +++ V+ YA G+PLAL+V+GS L++K WES ++ K +P+ +I ++LK+S+
Sbjct: 1099DPSYEDVLNRVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSF 1278
Query: 423 DGLDDEPKDIFLDITCFYAGDFVDKVVELL-DSCGFSADIGMDVLKDRGLISI-LEDRIA 480
D L +E K++FLDI C + G +V +L D G + VL ++ L+ + D +
Sbjct: 1279DALGEEQKNVFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSCCDTVE 1458
Query: 481 VHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIE 537
+HD++ +MGREI RQ+ +PGK L K+I V++ N GT I+ I LD + E
Sbjct: 1459MHDMIQDMGREIERQRSPEEPGKCKRLLLPKDIIQVLKDNTGTSKIEIICLDFSISDKEE 1638
Query: 538 KVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFP 597
V+ + F M N+++L ++ F +G + P GL+VL W +P P
Sbjct: 1639TVEWNENAFMKMKNLKILII-RNCKFSKGPN---------YFPEGLRVLEWHRYPSNCLP 1788
Query: 598 LDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQ---------- 647
+F P LV + D + HG S GHL V + +
Sbjct: 1789SNFDPINLVICKL---------PDSSITSFEFHGSSK-KLGHLTVLNFDRCEFLTKIPDV 1938
Query: 648 -ELPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLS 705
+LPNL+ L + LV + D + +K + GC+ LT P L LE LNL
Sbjct: 1939SDLPNLKELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSFP--PLNLTSLETLNLG 2112
Query: 706 GCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIG 765
GCS++E PEI M+N+ VL L I+ELP S +L+GL L L C + + S+
Sbjct: 2113GCSSLEYFPEILGEMKNITVLALHDLPIKELPFSFQNLIGLLFLWLDSCG-IVQLRCSLA 2289
Query: 766 TLTKLCKLDLTYC----------ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAE 815
T+ KLC+ +T E E GSI + T DC+ F I ++
Sbjct: 2290TMPKLCEFCITDSCNRWQWVESEEGEEKVVGSILSFEAT-----DCNLCDDFFFI--GSK 2448
Query: 816 SFAHI---SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGC 872
FAH+ +L LP + L L TL + +C L+ + NLK D C
Sbjct: 2449RFAHVGYLNLPGNNFTILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNLK---HFDARNC 2619
Query: 873 RKLT 876
LT
Sbjct: 2620ASLT 2631
Score = 86.3 bits (212), Expect = 7e-17
Identities = 96/356 (26%), Positives = 136/356 (37%), Gaps = 32/356 (8%)
Frame = +1
Query: 695 KLKFLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFC 754
KL L LN C + IP++ + L L+ELS +C
Sbjct: 1873 KLGHLTVLNFDRCEFLTKIPDVSD------------------------LPNLKELSFNWC 1980
Query: 755 SKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLD---LNDCSRLKTFPAIL 811
L ++ SIG L KL L C L +FP L LT L+ L CS L+ FP IL
Sbjct: 1981 ESLVAVDDSIGFLNKLKTLSAYGCRKLTSFP----PLNLTSLETLNLGGCSSLEYFPEIL 2148
Query: 812 EPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSEL---- 867
++ ++L + IK+LP S QNL+GL L L C + L S+ + L E
Sbjct: 2149 GEMKNITVLALHDLPIKELPFSFQNLIGLLFLWLDSC-GIVQLRCSLATMPKLCEFCITD 2325
Query: 868 DCS---------GCRKLTG-----------IPNDIGCLSSLRT-----LSLQDTGVVNLP 902
C+ G K+ G + +D + S R L+L LP
Sbjct: 2326 SCNRWQWVESEEGEEKVVGSILSFEATDCNLCDDFFFIGSKRFAHVGYLNLPGNNFTILP 2505
Query: 903 ESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIRSVMSNSRFLSDSKEGTFE 962
E L L +L V C+ L+ I LPP +K A +C S+ S S S L
Sbjct: 2506 EFFKELQFLTTLVVHDCKHLQEIRGLPPNLKHFDARNCASLTS-SSKSMLL--------- 2655
Query: 963 LYFTNNEKQDPGAQRDVVRNAEHRITDVACRFAFYCFPGSEVPNWFHHHCKRNSVT 1018
N E + G + FPG+ +P WF +S++
Sbjct: 2656 ----NQELHEAGGIE-------------------FVFPGTSIPEWFDQQSSGHSIS 2754
>TC225609 UP|Q84ZV3 (Q84ZV3) R 4 protein, complete
Length = 2688
Score = 451 bits (1161), Expect = e-127
Identities = 307/878 (34%), Positives = 462/878 (51%), Gaps = 47/878 (5%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNVF--IDDRSLETGDEIC----KAIEASTIYVI 77
YDVFLSF G DT FT +L+ AL + ++ IDD+ L GDEI KAI+ S I +
Sbjct: 34 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S+NYASS CLDELV I+ CK VIP+FY+VDPS VRHQ SYG+A +HQ R
Sbjct: 214 VLSQNYASSSFCLDELVTILHCKSE-GLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 390
Query: 138 -VPGVVMQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPP---FSQGMLG 192
+Q+W+ AL + ADLSGYH + E I I +++ +K++ + +G
Sbjct: 391 KAKKEKLQKWRMALKQVADLSGYHFEDGDAYEYKFIGSIVEEVSRKISRASLHVADYPVG 570
Query: 193 IDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQK 251
++ + ++ LL + S+ V IIGI GMGG+GK+TLA +Y+ + F C + N +++
Sbjct: 571 LESQVTEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALEVYNLIALHFDESCFLQNVREE 750
Query: 252 IDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
+++G+ L+ LSKLLGE DI G + R++R KVLLILDD+ + +
Sbjct: 751 SNKHGLKHLQSILLSKLLGEKDITLTSWQEGASTIQHRLQRKKVLLILDDVNKREQLKAI 930
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
+G FG GSR+I+T+R +H+LK E + YEV+ +++ +L+L NAFK + Y
Sbjct: 931 VGRPDWFGPGSRVIITTRDKHLLKCHEVERTYEVKVLNHNAALQLLTWNAFKREKIDPSY 1110
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLD 426
+++ V+ YA G+PLAL+++GS ++ K WES ++ K +PN +I ++LK+S+D L
Sbjct: 1111EDVLNRVVTYASGLPLALEIIGSNMFGKSVAGWESAVEHYKRIPNDEILEILKVSFDALG 1290
Query: 427 DEPKDIFLDITCFYAGDFVDKVVELL----DSCGFSADIGMDVLKDRGLISILEDRIAVH 482
+E K++FLDI G + +V +L D+C +DVL D+ LI + + +H
Sbjct: 1291EEQKNVFLDIAFCLKGCKLTEVEHMLCSLYDNC---MKHHIDVLVDKSLIKVKHGIVEMH 1461
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIEKV 539
DL+ +GREI RQ+ +PGK LW K+I HV++ N GT I+ I LD + E V
Sbjct: 1462DLIQVVGREIERQRSPEEPGKRKRLWLPKDIIHVLKDNTGTSKIEIICLDFSISYKEETV 1641
Query: 540 QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLD 599
+ + F M N+++L ++ F +G + P GL+VL W +P P +
Sbjct: 1642EFNENAFMKMENLKILII-RNGKFSKGPN---------YFPEGLRVLEWHRYPSNFLPSN 1791
Query: 600 FCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQ-----------E 648
F P LV + +S ++ HG S GHL V + +
Sbjct: 1792FDPINLVICKLPDSSIKSF---------EFHGSSK-KLGHLTVLKFDRCKFLTQIPDVSD 1941
Query: 649 LPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC 707
LPNL L + LV + D + +K++ GC+ LT P L LE L LS C
Sbjct: 1942LPNLRELSFEDCESLVAVDDSIGFLKKLKKLSAYGCRKLTSFP--PLNLTSLETLQLSSC 2115
Query: 708 SNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTL 767
S++E PEI MEN+ L L I+ELP S +L GL L+L C + +P S+ +
Sbjct: 2116SSLEYFPEILGEMENIRELRLTGLYIKELPFSFQNLTGLRLLALSGCG-IVQLPCSLAMM 2292
Query: 768 TKLCKLDLTYC--------ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAH 819
+L YC E E GSI K +C+ F L + FAH
Sbjct: 2293PELSSFYTDYCNRWQWIELEEGEEKLGSIISSKAQLFCATNCNLCDDF--FLAGFKRFAH 2466
Query: 820 I---SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESL 854
+ +L+ LP + L L+TL + +C L+ +
Sbjct: 2467VGYLNLSGNNFTILPEFFKELQFLRTLDVSDCEHLQEI 2580
Score = 92.0 bits (227), Expect = 1e-18
Identities = 99/326 (30%), Positives = 135/326 (41%), Gaps = 43/326 (13%)
Frame = +1
Query: 663 LVRIPDLSKSPNIKEIILSGCKSLT--RLPINLFKLKFLERLNLSGC----SNVENIP-- 714
++R SK PN G + L R P N F + +NL C S++++
Sbjct: 1690 IIRNGKFSKGPNY---FPEGLRVLEWHRYPSNFLPSNF-DPINLVICKLPDSSIKSFEFH 1857
Query: 715 EIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLD 774
+ + +L VL + ++ L L ELS + C L ++ SIG L KL KL
Sbjct: 1858 GSSKKLGHLTVLKFDRCKFLTQIPDVSDLPNLRELSFEDCESLVAVDDSIGFLKKLKKLS 2037
Query: 775 LTYCESLETFPGSIFDLKLTKLD---LNDCSRLKTFPAILEPAESFAHISLAETAIKKLP 831
C L +FP L LT L+ L+ CS L+ FP IL E+ + L IK+LP
Sbjct: 2038 AYGCRKLTSFP----PLNLTSLETLQLSSCSSLEYFPEILGEMENIRELRLTGLYIKELP 2205
Query: 832 SSLQNLVGLQTLCLKEC------------PDLESLPNSICNLKLLSELDCSGCRKLTGIP 879
S QNL GL+ L L C P+L S CN EL+ G KL I
Sbjct: 2206 FSFQNLTGLRLLALSGCGIVQLPCSLAMMPELSSFYTDYCNRWQWIELE-EGEEKLGSII 2382
Query: 880 NDIGCLSSLRTLSLQD------------TGVVNL--------PESIAHLSSLESLDVSYC 919
+ L +L D G +NL PE L L +LDVS C
Sbjct: 2383 SSKAQLFCATNCNLCDDFFLAGFKRFAHVGYLNLSGNNFTILPEFFKELQFLRTLDVSDC 2562
Query: 920 RKLECIPHLPPFMKQLLAFHCPSIRS 945
L+ I LPP ++ A +C S S
Sbjct: 2563 EHLQEIRGLPPILEYFDARNCVSFTS 2640
Score = 45.4 bits (106), Expect = 1e-04
Identities = 61/248 (24%), Positives = 97/248 (38%), Gaps = 27/248 (10%)
Frame = +1
Query: 720 MENLAVLVLKQTAIQELPSSLNHLVGLEELSL-KFCSKLESIPSSIGTLTK-LCKLDLTY 777
MENL +L+++ + P+ GL L ++ S +PS+ + +CKL +
Sbjct: 1669 MENLKILIIRNGKFSKGPNYFPE--GLRVLEWHRYPSNF--LPSNFDPINLVICKLPDSS 1836
Query: 778 CESLETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQN 836
+S E F GS L LT L + C L P + +
Sbjct: 1837 IKSFE-FHGSSKKLGHLTVLKFDRCKFLTQIPDV------------------------SD 1941
Query: 837 LVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTG------------------- 877
L L+ L ++C L ++ +SI LK L +L GCRKLT
Sbjct: 1942 LPNLRELSFEDCESLVAVDDSIGFLKKLKKLSAYGCRKLTSFPPLNLTSLETLQLSSCSS 2121
Query: 878 ---IPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCR--KLECIPHLPPFM 932
P +G + ++R L L + LP S +L+ L L +S C +L C + P +
Sbjct: 2122 LEYFPEILGEMENIRELRLTGLYIKELPFSFQNLTGLRLLALSGCGIVQLPCSLAMMPEL 2301
Query: 933 KQLLAFHC 940
+C
Sbjct: 2302 SSFYTDYC 2325
>TC225617 UP|Q84ZV8 (Q84ZV8) R 3 protein, complete
Length = 2697
Score = 448 bits (1152), Expect = e-126
Identities = 305/889 (34%), Positives = 467/889 (52%), Gaps = 42/889 (4%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFLSF G DT FT +L+ AL + + FIDD+ L GDEI KAI+ S I +
Sbjct: 34 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQESRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S+NYASS CLDELV ++ CK VIP+FY VDPS VR Q SYG+A +HQ R
Sbjct: 214 VLSQNYASSSFCLDELVTVLLCKRK-GLLVIPVFYNVDPSDVRQQKGSYGEAMAKHQKRF 390
Query: 138 -VPGVVMQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPP---FSQGMLG 192
+Q+W+ AL + ADLSGYH + E I I + + +++N + +G
Sbjct: 391 KAKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRTPLHVADYPVG 570
Query: 193 IDKHIAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQK 251
+ + +++ LL + S + V IIGI GMGG+GK+TLA A+Y+ + F C + N +++
Sbjct: 571 LGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALHFDESCFLQNVREE 750
Query: 252 IDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
+++G+ L+ LSKLLGE DI G + R++R KVLLILDD+ + +
Sbjct: 751 SNKHGLKHLQSIILSKLLGEKDINLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAI 930
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
+G FG GSR+I+T+R +H+LK E + YEV+ ++ +L+L + NAFK + Y
Sbjct: 931 VGRPDWFGPGSRVIITTRDKHILKYHEVERTYEVKVLNQSAALQLLKWNAFKREKNDPSY 1110
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLD 426
+++ V+ YA G+PLAL+++GS L+ K WES ++ K +P+ +I ++LK+S+D L
Sbjct: 1111EDVLNRVVTYASGLPLALEIIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFDALG 1290
Query: 427 DEPKDIFLDITCFYAGDFVDKVVELL----DSCGFSADIGMDVLKDRGLISILEDRIAVH 482
+E K++FLDI C G + +V +L D+C +DVL D+ L + + +H
Sbjct: 1291EEQKNVFLDIACCLKGCKLTEVEHMLRGLYDNC---MKHHIDVLVDKSLTKVRHGIVEMH 1461
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIEKV 539
DL+ +MGREI RQ+ +PGK LW+ K+I V++ N GT I+ I++D + E V
Sbjct: 1462DLIQDMGREIERQRSPEEPGKRKRLWSPKDIIQVLKHNTGTSKIEIIYVDFSISDKEETV 1641
Query: 540 QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLD 599
+ + F M N+++L ++ F +G + P GL+VL W +P P +
Sbjct: 1642EWNENAFMKMENLKILII-RNGKFSKGPN---------YFPQGLRVLEWHRYPSNCLPSN 1791
Query: 600 FCPEKLVKLHMRESRLE--QLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQELPNLERLDL 657
F P LV + +S + + G + L L L + + L +LPNL L
Sbjct: 1792FDPINLVICKLPDSSITSFEFHGSSKKLGHLT-VLKFDWCKFLTQIPDVSDLPNLRELSF 1968
Query: 658 SNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVENIPEI 716
LV + D + +K++ GC+ LT P L LE L LS CS++E PEI
Sbjct: 1969QWCESLVAVDDSIGFLKKLKKLNADGCRKLTSFP--PLNLTSLETLELSHCSSLEYFPEI 2142
Query: 717 KETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLT 776
MEN+ L L I+ELP S +L+GL++LS+ C + + S+ + KL
Sbjct: 2143LGEMENIERLDLHGLPIKELPFSFQNLIGLQQLSMFGCG-IVQLRCSLAMMPKLSAFKFV 2319
Query: 777 YC--------ESLETFPGSIFDLKLT----KLDLNDCSRLKTFPAILEPAESFAHI---S 821
C E E GSI + +C+ F L + FAH+ +
Sbjct: 2320NCNRWQWVESEEAEEKVGSIISSEARFWTHSFSAKNCNLCDDF--FLTGFKKFAHVGYLN 2493
Query: 822 LAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCS 870
L+ LP + L L +L + C L+ + NLK + +C+
Sbjct: 2494LSRNNFTILPEFFKELQFLGSLNVSHCKHLQEIRGIPQNLKQFNARNCA 2640
Score = 89.0 bits (219), Expect = 1e-17
Identities = 74/245 (30%), Positives = 108/245 (43%), Gaps = 39/245 (15%)
Frame = +1
Query: 740 LNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLD-- 797
++ L L ELS ++C L ++ SIG L KL KL+ C L +FP L LT L+
Sbjct: 1933 VSDLPNLRELSFQWCESLVAVDDSIGFLKKLKKLNADGCRKLTSFP----PLNLTSLETL 2100
Query: 798 -LNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPN 856
L+ CS L+ FP IL E+ + L IK+LP S QNL+GLQ L + C + L
Sbjct: 2101 ELSHCSSLEYFPEILGEMENIERLDLHGLPIKELPFSFQNLIGLQQLSMFGC-GIVQLRC 2277
Query: 857 SICNLKLLSELDCSGCRKLTGIPND-----IGCLSSLRT--------------------- 890
S+ + LS C + + ++ +G + S
Sbjct: 2278 SLAMMPKLSAFKFVNCNRWQWVESEEAEEKVGSIISSEARFWTHSFSAKNCNLCDDFFLT 2457
Query: 891 ----------LSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHC 940
L+L LPE L L SL+VS+C+ L+ I +P +KQ A +C
Sbjct: 2458 GFKKFAHVGYLNLSRNNFTILPEFFKELQFLGSLNVSHCKHLQEIRGIPQNLKQFNARNC 2637
Query: 941 PSIRS 945
S+ S
Sbjct: 2638 ASLTS 2652
>TC225612 homologue to UP|Q84ZU8 (Q84ZU8) R 10 protein, complete
Length = 2706
Score = 439 bits (1129), Expect = e-123
Identities = 309/901 (34%), Positives = 467/901 (51%), Gaps = 48/901 (5%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFL+F G DT FT +L+ AL K + F D++ L G+EI KAI+ S I +
Sbjct: 34 YDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLKAIQESRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S+NYASS CLDELV I+ CK VIP+FY VDPS VRHQ SYG +HQ R
Sbjct: 214 VLSKNYASSSFCLDELVTILHCKSE-GLLVIPVFYNVDPSDVRHQKGSYGVEMAKHQKRF 390
Query: 138 -VPGVVMQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPP---FSQGMLG 192
+Q+W+ AL + ADL GYH + E I I + + +++N + +G
Sbjct: 391 KAKKEKLQKWRIALKQVADLCGYHFKDGDAYEYKFIQSIVEQVSREINRAPLHVADYPVG 570
Query: 193 IDKHIAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQK 251
+ + +++ LL + S + V IIGI GMGG+GK+TLA A+++ + F C + N +++
Sbjct: 571 LGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNLIALHFDESCFLQNVREE 750
Query: 252 IDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
+++G+ L+ LSKLLGE DI G + R++R KVLLILDD+ + +
Sbjct: 751 SNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKREQLKAI 930
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
+G FG GSR+I+T+R +H+LK+ E + YEV+ ++ +L+L + NAFK + Y
Sbjct: 931 VGRPDWFGPGSRVIITTRDKHLLKHHEVERTYEVKVLNQSAALQLLKWNAFKREKIDPSY 1110
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLD 426
+++ V+ YA G+PLAL+V+GS L+ K WES ++ K +P+ +I ++LK+S+D L
Sbjct: 1111EDVLNRVVTYASGLPLALEVIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFDALG 1290
Query: 427 DEPKDIFLDITCFYAGDFVDKVVELLDSC-GFSADIGMDVLKDRGLISIL---EDRIAVH 482
+E K++FLDI C + G +V ++L + G + VL ++ LI + D + +H
Sbjct: 1291EEQKNVFLDIACCFRGYKWTEVDDILRALYGNCKKHHIGVLVEKSLIKLNCYGTDTVEMH 1470
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIEKV 539
DL+ +M REI R++ +PGK LW K+I V + N GT I+ I LD + E V
Sbjct: 1471DLIQDMAREIERKRSPQEPGKCKRLWLPKDIIQVFKDNTGTSKIEIICLDSSISDKEETV 1650
Query: 540 QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLD 599
+ + F M N+++L ++ F +G + P GL+VL W +P P +
Sbjct: 1651EWNENAFMKMENLKILII-RNDKFSKGPN---------YFPEGLRVLEWHRYPSNCLPSN 1800
Query: 600 FCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLH-----------QE 648
F P LV + +S + HG S FGHL V +
Sbjct: 1801FHPNNLVICKLPDSCMTSF---------EFHGPSK--FGHLTVLKFDNCKFLTQIPDVSD 1947
Query: 649 LPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRL-PINLFKLKFLERLNLSG 706
LPNL L LV + D + +K++ GC L P+NL L+ LE LS
Sbjct: 1948LPNLRELSFEECESLVAVDDSIGFLNKLKKLSAYGCSKLKSFPPLNLTSLQTLE---LSQ 2118
Query: 707 CSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGT 766
CS++E PEI MEN+ L L I+EL S +L+GL L+L+ C + +P S+
Sbjct: 2119CSSLEYFPEIIGEMENIKHLFLYGLPIKELSFSFQNLIGLRWLTLRSCG-IVKLPCSLAM 2295
Query: 767 LTKLCKLDLTYC--------ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFA 818
+ +L + + YC E E GSI K + DC+ F L +++FA
Sbjct: 2296MPELFEFHMEYCNRWQWVESEEGEKKVGSIPSSKAHRFSAKDCNLCDDF--FLTGSKTFA 2469
Query: 819 HI---SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKL 875
H+ +L LP + L L TL + +C L+ + NLK D C L
Sbjct: 2470HVGHLNLPGNNFTILPEFFKELQLLTTLMVNDCKHLQEIRGLPPNLK---HFDARNCASL 2640
Query: 876 T 876
T
Sbjct: 2641T 2643
Score = 91.3 bits (225), Expect = 2e-18
Identities = 74/245 (30%), Positives = 109/245 (44%), Gaps = 35/245 (14%)
Frame = +1
Query: 740 LNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKL--- 796
++ L L ELS + C L ++ SIG L KL KL C L++FP L LT L
Sbjct: 1939 VSDLPNLRELSFEECESLVAVDDSIGFLNKLKKLSAYGCSKLKSFP----PLNLTSLQTL 2106
Query: 797 DLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPN 856
+L+ CS L+ FP I+ E+ H+ L IK+L S QNL+GL+ L L+ C + LP
Sbjct: 2107 ELSQCSSLEYFPEIIGEMENIKHLFLYGLPIKELSFSFQNLIGLRWLTLRSC-GIVKLPC 2283
Query: 857 SICNLKLLSELDCSGCRKLTGIPND-----IGCLSSLRT--------------------- 890
S+ + L E C + + ++ +G + S +
Sbjct: 2284 SLAMMPELFEFHMEYCNRWQWVESEEGEKKVGSIPSSKAHRFSAKDCNLCDDFFLTGSKT 2463
Query: 891 ------LSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIR 944
L+L LPE L L +L V+ C+ L+ I LPP +K A +C S+
Sbjct: 2464 FAHVGHLNLPGNNFTILPEFFKELQLLTTLMVNDCKHLQEIRGLPPNLKHFDARNCASLT 2643
Query: 945 SVMSN 949
S N
Sbjct: 2644 SSSKN 2658
>TC225614 UP|Q84ZV7 (Q84ZV7) R 12 protein, partial (88%)
Length = 2367
Score = 433 bits (1114), Expect = e-121
Identities = 286/795 (35%), Positives = 429/795 (52%), Gaps = 35/795 (4%)
Frame = +1
Query: 21 PLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTI 74
P YDVFLSF EDT + FT +L+ AL + + F D+ L E+ KAI AS +
Sbjct: 25 PFIYDVFLSFIREDTHRGFTFYLYKALNDRGIYTFFYDQELPRETEVTPGLYKAILASRV 204
Query: 75 YVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQ 134
+IV SENYA S CLDELV I+ C +R VIP+F+ VDPS VRHQ SYG+A +HQ
Sbjct: 205 AIIVLSENYAFSSFCLDELVTILHC----EREVIPVFHNVDPSDVRHQKGSYGEAMAKHQ 372
Query: 135 NRLVPGVVMQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDI---FKKLNPPFSQGM 190
R +Q+W+ AL + A+L GYH S E MLI RI + + F + +
Sbjct: 373 KRF-KAKKLQKWRMALKQVANLCGYHFKDGGSYEYMLIGRIVKQVSRMFGLASLHVADYP 549
Query: 191 LGIDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQ 249
+G++ + ++ LL + S+ V IIGI GMGG+GK+TLA A+Y+ + F C + N +
Sbjct: 550 VGLESQVTEVMKLLDVGSDDVVHIIGIHGMGGLGKTTLAMAVYNFIAPHFDESCFLQNVR 729
Query: 250 QKIDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPIL 304
++ +++G+ L+ LSKLLGE DI G + R++ K+LLILDD+ +
Sbjct: 730 EESNKHGLKHLQSVLLSKLLGEKDITLTSWQEGASMIQHRLRLKKILLILDDVDKREQLK 909
Query: 305 ELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLE 364
++G FG GSR+I+T+R +H+LK + + YEV +++ D+L+L NAFK +
Sbjct: 910 AIVGKPDWFGPGSRVIITTRDKHLLKYHDVERTYEVNVLNHDDALQLLTWNAFKREKIDP 1089
Query: 365 GYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDG 424
Y +++ + YA G+PLAL+V+GS LY K WES L+ K +P+ +I +L++S+D
Sbjct: 1090SYKDVLNRAVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILQVSFDA 1269
Query: 425 LDDEPKDIFLDITCFYAG----DFVDKVVELLDSCGFSADIGMDVLKDRGLISILEDRIA 480
L++E K++FLDI C + G + D L +C IG+ V K L D +
Sbjct: 1270LEEEQKNVFLDIACCFKGYKWTEVYDIFRALYSNCKMH-HIGVLVEKSLLLKVSWRDNVE 1446
Query: 481 VHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIE 537
+HDL+ +MGR+I RQ+ +PGK LW+ K+I V++ N GT ++ I LD + E
Sbjct: 1447MHDLIQDMGRDIERQRSPEEPGKCKRLWSPKDIIQVLKHNTGTSKLEIICLDSSISDKEE 1626
Query: 538 KVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFP 597
V+ + F M N+++L ++ F +G + P GL+VL W +P P
Sbjct: 1627TVEWNENAFMKMENLKILII-RNGKFSKGPN---------YFPEGLRVLEWHRYPSNCLP 1776
Query: 598 LDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQ---------- 647
+F P LV + +S + L HG S + GHL V +
Sbjct: 1777SNFDPINLVICKLPDSSITSL---------EFHGSSKL--GHLTVLKFDKCKFLTQIPDV 1923
Query: 648 -ELPNLERLDLSNSWKLVRIPDLSKSPNIKEII-LSGCKSLTRLPINLFKLKFLERLNLS 705
+LPNL L LV I D N EI+ +GC+ LT P L LE L LS
Sbjct: 1924SDLPNLRELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLTSFP--PLNLTSLETLELS 2097
Query: 706 GCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIG 765
CS++E PEI MEN+ L L++ I+ELP S +L+GL E++L+ C ++ + S+
Sbjct: 2098HCSSLEYFPEILGEMENITALHLERLPIKELPFSFQNLIGLREITLRRC-RIVRLRCSLA 2274
Query: 766 TLTKLCKLDLTYCES 780
+ L + + C S
Sbjct: 2275MMPNLFRFQIRNCNS 2319
Score = 70.9 bits (172), Expect = 3e-12
Identities = 45/112 (40%), Positives = 60/112 (53%), Gaps = 3/112 (2%)
Frame = +1
Query: 740 LNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLD-- 797
++ L L ELS C L +I SIG L KL L+ C L +FP L LT L+
Sbjct: 1921 VSDLPNLRELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLTSFP----PLNLTSLETL 2088
Query: 798 -LNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKEC 848
L+ CS L+ FP IL E+ + L IK+LP S QNL+GL+ + L+ C
Sbjct: 2089 ELSHCSSLEYFPEILGEMENITALHLERLPIKELPFSFQNLIGLREITLRRC 2244
Score = 47.8 bits (112), Expect = 3e-05
Identities = 56/221 (25%), Positives = 85/221 (38%), Gaps = 1/221 (0%)
Frame = +1
Query: 720 MENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTK-LCKLDLTYC 778
MENL +L+++ + P+ GL L +PS+ + +CKL +
Sbjct: 1660 MENLKILIIRNGKFSKGPNYFPE--GLRVLEWHRYPS-NCLPSNFDPINLVICKLPDSSI 1830
Query: 779 ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLV 838
SLE F GS LT L + C L P + +L
Sbjct: 1831 TSLE-FHGSSKLGHLTVLKFDKCKFLTQIPDV------------------------SDLP 1935
Query: 839 GLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGV 898
L+ L C L ++ +SI L L L+ +GCRKLT P
Sbjct: 1936 NLRELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLTSFPP------------------ 2061
Query: 899 VNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFH 939
+L+SLE+L++S+C LE P + M+ + A H
Sbjct: 2062 -------LNLTSLETLELSHCSSLEYFPEILGEMENITALH 2163
>TC225615 UP|Q84ZU7 (Q84ZU7) R 5 protein, complete
Length = 2724
Score = 433 bits (1114), Expect = e-121
Identities = 301/894 (33%), Positives = 462/894 (51%), Gaps = 47/894 (5%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFLSF GEDT FT +L+ AL K + F D+ L +G+EI KAI+ S I +
Sbjct: 34 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V SE++ASS CLDEL I+ C VIP+FY+V P VRHQ +YG+A +H+ R
Sbjct: 214 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 393
Query: 138 VPGVVMQRWKKALLEAADLSGYH-SIAARSESMLIDRIAQDIFKKLNPP---FSQGMLGI 193
+Q+W++AL + A+LSG H E I RI + +K+NP + +G+
Sbjct: 394 PDK--LQKWERALRQVANLSGLHFKDRDEYEYKFIGRIVASVSEKINPASLHVADLPVGL 567
Query: 194 DKHIAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEALYHKL--GTQFSSRCLIVNAQQ 250
+ + +++ LL + + + V +IGI GMGGIGKSTLA A+Y+ L F C + N ++
Sbjct: 568 ESKVQEVRKLLDVGNHDGVCMIGIHGMGGIGKSTLARAVYNDLIITENFDGLCFLENVRE 747
Query: 251 KIDRYGIYSLRKKYLSKLLGEDIQSNGLNYAIERV----KRAKVLLILDDLKISIPILEL 306
+ +G+ L+ LS++LGEDI+ I ++ K KVLLILDD+ + +
Sbjct: 748 SSNNHGLQHLQSILLSEILGEDIKVRSKQQGISKIQSMLKGKKVLLILDDVDKPQQLQTI 927
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
G FG GS II+T+R + +L + YEV+ ++ +L+L NAFK + Y
Sbjct: 928 AGRRDWFGPGSIIIITTRDKQLLAPHGVKKRYEVEVLNQNAALQLLTWNAFKREKIDPSY 1107
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLD 426
+++ V+ YA G+PLAL+V+GS ++ K W+S ++ K +PN +I ++LK+S+D L
Sbjct: 1108EDVLNRVVTYASGLPLALEVIGSNMFGKRVAEWKSAVEHYKRIPNDEILEILKVSFDALG 1287
Query: 427 DEPKDIFLDITCFYAGDFVDKVVELL----DSCGFSADIGMDVLKDRGLISILEDRIAVH 482
+E K++FLDI C + G + +V +L ++C +DVL D+ LI + + +H
Sbjct: 1288EEQKNVFLDIACCFKGCKLTEVEHMLRGLYNNC---MKHHIDVLVDKSLIKVRHGTVNMH 1458
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEK---V 539
DL+ +GREI RQ +PGK LW K+I V++ N GT I+ I LD +K V
Sbjct: 1459DLIQVVGREIERQISPEEPGKCKRLWLPKDIIQVLKHNTGTSKIEIICLDFSISDKEQTV 1638
Query: 540 QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLD 599
+ + F M N+++L ++ F +G + P GL+VL W +P + P +
Sbjct: 1639EWNQNAFMKMENLKILII-RNGKFSKGPN---------YFPEGLRVLEWHRYPSKCLPSN 1788
Query: 600 FCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLH-----------QE 648
F P L+ + +S + HG S FGHL V +
Sbjct: 1789FHPNNLLICKLPDSSMASF---------EFHGSSK--FGHLTVLKFDNCKFLTQIPDVSD 1935
Query: 649 LPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC 707
LPNL L LV + D + +K++ GC+ LT P L LE L LSGC
Sbjct: 1936LPNLRELSFKGCESLVAVDDSIGFLNKLKKLNAYGCRKLTSFP--PLNLTSLETLQLSGC 2109
Query: 708 SNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTL 767
S++E PEI MEN+ LVL+ I+EL S +L+GL+ L L C +E +P + +
Sbjct: 2110SSLEYFPEILGEMENIKQLVLRDLPIKELQFSFQNLIGLQVLYLWSCLIVE-LPCRLAMM 2286
Query: 768 TKLCKLDLTYC--------ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAH 819
+L +L + YC E E GSI K +C+ F L +++F H
Sbjct: 2287PELFQLHIEYCNRWQWVESEEGEEKVGSILSSKARWFRAMNCNLCDDF--FLTGSKTFTH 2460
Query: 820 I---SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCS 870
+ L+ LP + L L+TL + +C L+ + NLK ++C+
Sbjct: 2461VEYLDLSGNNFTILPEFFKELKFLRTLDVSDCEHLQKIRGLPPNLKDFRAINCA 2622
Score = 95.9 bits (237), Expect = 9e-20
Identities = 79/241 (32%), Positives = 107/241 (43%), Gaps = 35/241 (14%)
Frame = +1
Query: 740 LNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLD-- 797
++ L L ELS K C L ++ SIG L KL KL+ C L +FP L LT L+
Sbjct: 1927 VSDLPNLRELSFKGCESLVAVDDSIGFLNKLKKLNAYGCRKLTSFP----PLNLTSLETL 2094
Query: 798 -LNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPN 856
L+ CS L+ FP IL E+ + L + IK+L S QNL+GLQ L L C +E LP
Sbjct: 2095 QLSGCSSLEYFPEILGEMENIKQLVLRDLPIKELQFSFQNLIGLQVLYLWSCLIVE-LPC 2271
Query: 857 SICNLKLLSELDCSGCRKLTGIPNDIG------CLSS----------------------- 887
+ + L +L C + + ++ G LSS
Sbjct: 2272 RLAMMPELFQLHIEYCNRWQWVESEEGEEKVGSILSSKARWFRAMNCNLCDDFFLTGSKT 2451
Query: 888 ---LRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIR 944
+ L L LPE L L +LDVS C L+ I LPP +K A +C S+
Sbjct: 2452 FTHVEYLDLSGNNFTILPEFFKELKFLRTLDVSDCEHLQKIRGLPPNLKDFRAINCASLT 2631
Query: 945 S 945
S
Sbjct: 2632 S 2634
>TC225611 homologue to UP|Q84ZU6 (Q84ZU6) R 1 protein, complete
Length = 2709
Score = 427 bits (1098), Expect = e-119
Identities = 301/900 (33%), Positives = 461/900 (50%), Gaps = 47/900 (5%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFL+F GEDT FT +L+ AL K + F D+ L +GD+I KAI+ S I +
Sbjct: 34 YDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSKAIQESRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S+NYASS CLDELV I+ CK VIP+F+ VDPS VRH SYG+A +HQ R
Sbjct: 214 VLSQNYASSSFCLDELVTILHCKRE-GLLVIPVFHNVDPSAVRHLKGSYGEAMAKHQKRF 390
Query: 138 -VPGVVMQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPP---FSQGMLG 192
+Q+W+ AL + ADLSGYH + E I I +++ +K+N + +G
Sbjct: 391 KAKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIGNIVEEVSRKINCAPLHVADYPVG 570
Query: 193 IDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQK 251
+ + ++ LL + S+ V IIGI GMGG+GK+TLA A+Y+ F C + N +++
Sbjct: 571 LGSQVIEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALAVYNIFALYFDESCFLQNVREE 750
Query: 252 IDRYGIYSLRKKYLSKLLGE-DI----QSNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
++ + L+ LSKLLGE DI G + R++R KVLLILDD+ ++ L
Sbjct: 751 SKKHELKHLQSIVLSKLLGEKDIILTSSQEGASMIQHRLRRKKVLLILDDVDKREQLMAL 930
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
+G FG GSR+I+T+R +H+LK + + YEV+ +++ +L+L NAFK + Y
Sbjct: 931 VGRSDWFGPGSRVIITTRDKHLLKYHDVERTYEVKVLNHNAALQLLTWNAFKREKIDPIY 1110
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLD 426
+++ V+ YA G+PLAL+V+GS L+ K WES ++ K +P+ +I +LK+S+D L
Sbjct: 1111DDVLNRVVTYASGLPLALEVIGSDLFGKTVAEWESAVEHYKRIPSDEILKILKVSFDALG 1290
Query: 427 DEPKDIFLDITCFYAGDFVDKVVELLDSC-GFSADIGMDVLKDRGLISIL---EDRIAVH 482
+E K++FLDI C + G +V ++L + G + L ++ LI + + +H
Sbjct: 1291EEQKNVFLDIACCFKGYKWKEVDDILRALYGNCKKHHIGELVEKSLIKLNCYDSGTVEMH 1470
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIEKV 539
DL+ +MGREI RQ+ +P K LW+ K+I V++ N GT I+ I LD + E V
Sbjct: 1471DLIQDMGREIERQRSPEEPWKCKRLWSPKDIIQVLKHNTGTSKIEIICLDFSISDKEETV 1650
Query: 540 QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLD 599
+ + F M N+++L ++ F +G + P GL VL W +P P +
Sbjct: 1651EWNENAFMKMENLKILII-RNGKFSKGPN---------YFPEGLTVLEWHRYPSNCLPYN 1800
Query: 600 FCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFFGHLIVFSLHQ-----------E 648
F P L+ + D + HG S F HL V + Q +
Sbjct: 1801FHPNNLLICKL---------PDSSITSFELHGPSK--FWHLTVLNFDQCEFLTQIPDVSD 1947
Query: 649 LPNLERLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGC 707
LPNL+ L L+ + D + +K++ GC+ L P L LE L LSGC
Sbjct: 1948LPNLKELSFDWCESLIAVDDSIGFLNKLKKLSAYGCRKLRSFP--PLNLTSLETLQLSGC 2121
Query: 708 SNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTL 767
S++E PEI MEN+ L L I+ELP S +L+GL L+L C ++ +P S+ +
Sbjct: 2122SSLEYFPEILGEMENIKALDLDGLPIKELPFSFQNLIGLCRLTLNSCGIIQ-LPCSLAMM 2298
Query: 768 TKLCKLDLTYC--------ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFA- 818
+L + C E E GSI K +C+ F L ++ F
Sbjct: 2299PELSVFRIENCNRWHWVESEEGEEKVGSITSSKKLSFTAMNCNLCDDF--FLTGSKRFTR 2472
Query: 819 --HISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
++ L+ LP + L L+ L + +C L+ + NL+ +C+ T
Sbjct: 2473VEYLDLSGNNFTILPEFFKELKFLRALMVSDCEHLQEIRGLPPNLEYFHARNCASLTSST 2652
Score = 91.3 bits (225), Expect = 2e-18
Identities = 79/259 (30%), Positives = 112/259 (42%), Gaps = 35/259 (13%)
Frame = +1
Query: 722 NLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESL 781
+L VL Q ++ L L+ELS +C L ++ SIG L KL KL C L
Sbjct: 1885 HLTVLNFDQCEFLTQIPDVSDLPNLKELSFDWCESLIAVDDSIGFLNKLKKLSAYGCRKL 2064
Query: 782 ETFPGSIFDLKLTKLD---LNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLV 838
+FP L LT L+ L+ CS L+ FP IL E+ + L IK+LP S QNL+
Sbjct: 2065 RSFP----PLNLTSLETLQLSGCSSLEYFPEILGEMENIKALDLDGLPIKELPFSFQNLI 2232
Query: 839 GLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPND-----IGCLSSLRTLS- 892
GL L L C ++ LP S+ + LS C + + ++ +G ++S + LS
Sbjct: 2233 GLCRLTLNSCGIIQ-LPCSLAMMPELSVFRIENCNRWHWVESEEGEEKVGSITSSKKLSF 2409
Query: 893 --------------------------LQDTGVVNLPESIAHLSSLESLDVSYCRKLECIP 926
L LPE L L +L VS C L+ I
Sbjct: 2410 TAMNCNLCDDFFLTGSKRFTRVEYLDLSGNNFTILPEFFKELKFLRALMVSDCEHLQEIR 2589
Query: 927 HLPPFMKQLLAFHCPSIRS 945
LPP ++ A +C S+ S
Sbjct: 2590 GLPPNLEYFHARNCASLTS 2646
>TC225613 UP|Q9FVK5 (Q9FVK5) Resistance protein LM6 (Fragment), partial (98%)
Length = 2597
Score = 422 bits (1086), Expect = e-118
Identities = 308/897 (34%), Positives = 461/897 (51%), Gaps = 52/897 (5%)
Frame = +1
Query: 32 GEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEI----CKAIEASTIYVIVFSENYAS 85
G+DT + FT +L+ AL + + FIDD+ L GDEI AI+ S I + V S+NYAS
Sbjct: 7 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 186
Query: 86 SRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRLVPGVV-MQ 144
S CLDELV I+ CK + VIP+FY+VDPS VRHQ SYG+A +HQ R +Q
Sbjct: 187 SSFCLDELVTILHCK-SQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFKANKEKLQ 363
Query: 145 RWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPPFSQGML-------GIDKH 196
+W+ AL + ADLSGYH S E I I ++I +K FS+ L G++
Sbjct: 364 KWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRK----FSRASLHVADYPVGLESE 531
Query: 197 IAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRY 255
+ ++ LL + S + V IIGI GMGG+GK+TLA A+++ + F C + N +++ +++
Sbjct: 532 VTEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALHFDESCFLQNVREESNKH 711
Query: 256 GIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPILELIGGH 310
G+ L+ LSKLLGE DI G + R++R KVLLILDD+ + ++G
Sbjct: 712 GLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRP 891
Query: 311 GNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLI 370
FG GSR+I+T+R +H+LK E + YEV+ ++ +L+L NAFK + Y +++
Sbjct: 892 DWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLTWNAFKREKIDPSYEDVL 1071
Query: 371 VEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDDEPK 430
V+ YA G+PLAL+V+GS L++K WES ++ K +P+ +I ++LK+S+D L +E K
Sbjct: 1072NRVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSFDALGEEQK 1251
Query: 431 DIFLDITCFYAGDFVDKVVELL-DSCGFSADIGMDVLKDRGLISI-LEDRIAVHDLVLEM 488
++FLDI C + G +V +L D G + VL ++ L+ + D + +HD++ +M
Sbjct: 1252NVFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSCCDTVEMHDMIQDM 1431
Query: 489 GREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMC---QIEKVQLHPEI 545
GREI RQ+ +PGK L K+I V + I+ I LD + E V+ +
Sbjct: 1432GREIERQRSPEEPGKCKRLLLPKDIIQVFK-------IEIICLDFSISDKEETVEWNENA 1590
Query: 546 FKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKL 605
F M N+++L ++ F +G + P GL+VL W +P P +F P L
Sbjct: 1591FMKMKNLKILII-RNCKFSKGPN---------YFPEGLRVLEWHRYPSNCLPSNFDPINL 1740
Query: 606 VKLHMRESRLEQL-------WGDDQVLQSLCHGLSYIFFG-----HLIVFSLHQELPNLE 653
V + +S + D + HG S L +LPNL+
Sbjct: 1741VICKLPDSSITSFEFLVICKLPDSSITSFEFHGSSKAILNFDRCEFLTKIPDVSDLPNLK 1920
Query: 654 RLDLSNSWKLVRIPD-LSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNLSGCSNVEN 712
L + LV + D + +K + GC+ LT P L LE LNL GCS++E
Sbjct: 1921ELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSFP--PLNLTSLETLNLGGCSSLEY 2094
Query: 713 IPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCK 772
PEI M+N+ VL L I+ELP S +L+GL L L C + + S+ T+ KLC+
Sbjct: 2095FPEILGEMKNITVLALHDLPIKELPFSFQNLIGLLFLWLDSCG-IVQLRCSLATMPKLCE 2271
Query: 773 LDLTYC----------ESLETFPGSIFDLKLTKLDLNDCSRLKTFPAILEPAESFAHI-- 820
+T E E GSI + T DC+ F L ++ FAH+
Sbjct: 2272FCITDSCNRWQWVESEEGEEKVVGSILSFEAT-----DCNLCDDF--FLIGSKRFAHVGY 2430
Query: 821 -SLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLT 876
+L LP + L L TL + +C L+ + NLK D C LT
Sbjct: 2431LNLPGNNFTILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNLK---HFDARNCASLT 2592
Score = 81.6 bits (200), Expect = 2e-15
Identities = 75/238 (31%), Positives = 106/238 (44%), Gaps = 32/238 (13%)
Frame = +1
Query: 740 LNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLD-- 797
++ L L+ELS +C L ++ SIG L KL L C L +FP L LT L+
Sbjct: 1897 VSDLPNLKELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSFP----PLNLTSLETL 2064
Query: 798 -LNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNLVGLQTLCLKECPDLESLPN 856
L CS L+ FP IL ++ ++L + IK+LP S QNL+GL L L C + L
Sbjct: 2065 NLGGCSSLEYFPEILGEMKNITVLALHDLPIKELPFSFQNLIGLLFLWLDSC-GIVQLRC 2241
Query: 857 SICNLKLLSEL----DCS---------GCRKLTG-----------IPNDIGCLSSLRT-- 890
S+ + L E C+ G K+ G + +D + S R
Sbjct: 2242 SLATMPKLCEFCITDSCNRWQWVESEEGEEKVVGSILSFEATDCNLCDDFFLIGSKRFAH 2421
Query: 891 ---LSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIRS 945
L+L LPE L L +L V C+ L+ I LPP +K A +C S+ S
Sbjct: 2422 VGYLNLPGNNFTILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNLKHFDARNCASLTS 2595
>TC207788 homologue to UP|Q8W2C0 (Q8W2C0) Functional candidate resistance
protein KR1, complete
Length = 3949
Score = 412 bits (1059), Expect = e-115
Identities = 345/1191 (28%), Positives = 559/1191 (45%), Gaps = 39/1191 (3%)
Frame = +1
Query: 1 MSESSSSSSAAAAAMAPPPNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRS 58
M++ SSSSS + DVFLSF GEDT + FT +L+ AL + + F+DD+
Sbjct: 100 MAKQSSSSSFSYR--------FSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKK 255
Query: 59 LETGDEIC----KAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRV 114
+ GD+I KAIE S I++IV SENYASS CL+EL I++ ++P+FY+V
Sbjct: 256 IPRGDQITSGLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKV 435
Query: 115 DPSTVRHQTNSYGDAFDEHQNRLVPGVVMQR---WKKALLEAADLSGYHSI--AARSESM 169
DPS VR+ T S+G A H+ + M++ WK AL + A+LSGYH E
Sbjct: 436 DPSDVRNHTGSFGKALTNHEKKFKSTNDMEKLETWKMALNKVANLSGYHHFKHGEEYEYE 615
Query: 170 LIDRIAQDIFKKLNPP---FSQGMLGIDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKS 225
I RI + + KK+N + +G++ I ++++LL + S+ V ++GI G+GG+GK+
Sbjct: 616 FIQRIVELVSKKINRAPLHVADYPVGLESRIQEVKALLDVGSDDVVHMLGIHGLGGVGKT 795
Query: 226 TLAEALYHKLGTQFSSRCLIVNAQQKIDRYGIYSLRKKYLSKLLGEDIQ---SNGLNYAI 282
TLA A+Y+ + F + C + N ++ ++GI L++ LS+ GE G++
Sbjct: 796 TLAAAVYNSIADHFEALCFLQNVRETSKKHGIQHLQRNLLSETAGEHKLIGVKQGISIIQ 975
Query: 283 ERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQG 342
R+++ K+LLILDD+ + L G FG GSR+I+T+R + +L + YEV
Sbjct: 976 HRLQQKKILLILDDVDKREQLQALAGRPDLFGPGSRVIITTRDKQLLACHGVERTYEVNE 1155
Query: 343 MSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESE 402
++ + +LEL AFK + Y +++ YA G+PLAL+V+GS L K E W S
Sbjct: 1156 LNEEYALELLNWKAFKLEKVDPFYKDVLNRAATYASGLPLALEVIGSNLSGKNIEQWISA 1335
Query: 403 LKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELLDS-CGFSADI 461
L + K +PN +I ++LK+SYD L+++ + IFLDI C + + ++ ++L + G
Sbjct: 1336 LDRYKRIPNKEIQEILKVSYDALEEDEQSIFLDIACCFKKYDLAEIQDILHAHHGHCMKH 1515
Query: 462 GMDVLKDRGLISI-LEDRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKN 520
+ VL ++ LI I L+ + +HDL+ +MG+EIVR++ +PGK S LW +I V+ +N
Sbjct: 1516 HIGVLVEKSLIKISLDGYVTLHDLIEDMGKEIVRKESPQEPGKRSRLWLPTDIVQVLEEN 1695
Query: 521 KGTDAIQCI---FLDMCQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLE 577
KGT I I F + ++Q + FK M N++ L + F +G +
Sbjct: 1696 KGTSHIGIICMNFYSSFEEVEIQWDGDAFKKMKNLKTLII-RSGHFSKGPKH-------- 1848
Query: 578 SLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQSLCHGLSYIFF 637
P L+VL W +P FP DF EKL ++ + F
Sbjct: 1849 -FPKSLRVLEWWRYPSHYFPYDFQMEKLAIFNLPDCG---------------------FT 1962
Query: 638 GHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLK 697
+ L ++ NL L+ + L IPD+S P+++++ C +L + ++ L+
Sbjct: 1963 SRELAAMLKKKFVNLTSLNFDSCQHLTLIPDVSCVPHLQKLSFKDCDNLYAIHPSVGFLE 2142
Query: 698 FLERLNLSGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKL 757
L L+ GCS ++N P IK L LE+L L FC L
Sbjct: 2143 KLRILDAEGCSRLKNFPPIK-------------------------LTSLEQLKLGFCHSL 2247
Query: 758 ESIPSSIGTLTKLCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLKT----FPAILEP 813
E+ P +G + + +LD LE P F L L +RL+T FP
Sbjct: 2248 ENFPEILGKMENITELD------LEQTPVKKFTLSFQNL-----TRLQTVLLCFPR--NQ 2388
Query: 814 AESFAHISLAETAIKKLPSSLQNLVGL---QTLCLKECPDLESLP-NSICNLKLLSELDC 869
I L+ + L N++G+ L KE E++ + N++ L +C
Sbjct: 2389 TNGCTGIFLSNICPMQESPELINVIGVGWEGCLFRKEDEGAENVSLTTSSNVQFLDLRNC 2568
Query: 870 SGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPESIAHLSSLESLDVSYCRKLECIPHLP 929
+ I + C +++ L+L +PE I L LD++YC +L I +P
Sbjct: 2569 NLSDDFFRIA--LPCFANVMRLNLSRNNFTVIPECIKECRFLTMLDLNYCERLREIRGIP 2742
Query: 930 PFMKQLLAFHCPSIRSVMSNSRFLSDSKEGTFELYFTNNEKQDPGAQRDVVRNAEHRITD 989
P +K A C S+ S S LS E + G
Sbjct: 2743 PNLKYFYAEECLSLTS-SCRSMLLS-------------QELHEAGR-------------- 2838
Query: 990 VACRFAFYCFPGSEVPNWFHHHCKRNSVTVDKDSLNLSNDNRIIGFAMCFVLQ--LEDMN 1047
F+ PG+++P WF + ++ N+ A+C +++ E +
Sbjct: 2839 -----TFFYLPGAKIPEWF-------DFQTSEFPISFWFRNKFPAIAICHIIKRVAEFSS 2982
Query: 1048 YTGYRF-SNFRCRLTFESDGQTHILPIDDQLNNSLHQIDLRVGHLVRDHTFIWNYYLMDS 1106
G+ F N R ++ +G ++ ++ DLR G V D+ + L+++
Sbjct: 2983 SRGWTFRPNIRTKVII--NGNANLFNSVVLGSDCTCLFDLR-GERVTDNL---DEALLEN 3144
Query: 1107 ASNHNGI-IHAHNFTFE---ISNGLY-LKSSSLVKECGIYPLYRKEKDDDD 1152
NH + FTF I GL+ LK S +++ RK K D+D
Sbjct: 3145 EWNHAEVTCPGFTFTFAPTFIKTGLHVLKQESNMEDIRFSDPCRKTKLDND 3297
>TC228427 UP|Q8H6S7 (Q8H6S7) Resistance protein KR3, complete
Length = 2358
Score = 352 bits (904), Expect = 4e-97
Identities = 216/607 (35%), Positives = 339/607 (55%), Gaps = 29/607 (4%)
Frame = +1
Query: 23 KYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYV 76
+YDVF++F GEDT FT HLH AL K + F+D+ ++ GDEI +AI+ S I +
Sbjct: 190 RYDVFINFRGEDTRFAFTGHLHKALCNKGIRAFMDENDIKRGDEIRATLEEAIKGSRIAI 369
Query: 77 IVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNR 136
VFS++YASS CLDEL I+ C VIP+FY+VDPS VR SY + + R
Sbjct: 370 TVFSKDYASSSFCLDELATILGCYREKTLLVIPVFYKVDPSDVRRLQGSYAEGLARLEER 549
Query: 137 LVPGVVMQRWKKALLEAADLSGYH-SIAARSESMLIDRIAQDIFKKLNPP-----FSQGM 190
P M+ WKKAL + A+L+G+H A E I +I D+F K+N +
Sbjct: 550 FHPN--MENWKKALQKVAELAGHHFKDGAGYEFKFIRKIVDDVFDKINKAEASIYVADHP 723
Query: 191 LGIDKHIAQIQSLLQL-DSEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQ 249
+G+ + +I+ LL+ S+A+ +IGI GMGG+GKSTLA A+Y+ F C + N +
Sbjct: 724 VGLHLEVEKIRKLLEAGSSDAISMIGIHGMGGVGKSTLARAVYNLHTDHFDDSCFLQNVR 903
Query: 250 QKIDRYGIYSLRKKYLSKLLGEDI----QSNGLNYAIERVKRAKVLLILDDLKISIPILE 305
++ +R+G+ L+ LS++L ++I + G + ++K KVLL+LDD+ +
Sbjct: 904 EESNRHGLKRLQSILLSQILKKEINLASEQQGTSMIKNKLKGKKVLLVLDDVDEHKQLQA 1083
Query: 306 LIG----GHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKE-D 360
++G FG +I+T+R + +L + +EV+ +S +D+++L + AFK D
Sbjct: 1084IVGKSVWSESEFGTRLVLIITTRDKQLLTSYGVKRTHEVKELSKKDAIQLLKRKAFKTYD 1263
Query: 361 YPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKL 420
+ Y ++ +V+ + G+PLAL+V+GS L+ K + WES +K+ + +PN +I +LK+
Sbjct: 1264EVDQSYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEILKILKV 1443
Query: 421 SYDGLDDEPKDIFLDITCFYAG----DFVDKVVELLDSCGFSADIGMDVLKDRGLISILE 476
S+D L++E K +FLDITC G + D + L D+C IG VL D+ LI I +
Sbjct: 1444SFDALEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNC-MKYHIG--VLVDKSLIQISD 1614
Query: 477 DRIAVHDLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDM--- 533
DR+ +HDL+ MG+EI RQ+ + GK LW K+I V++ N GT ++ I LD
Sbjct: 1615DRVTLHDLIENMGKEIDRQKSPKETGKRRRLWLLKDIIQVLKDNSGTSEVKIICLDFPIS 1794
Query: 534 CQIEKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQ 593
+ E ++ + FK M N++ L ++ QG + LP L++L W P
Sbjct: 1795DKQETIEWNGNAFKEMKNLKALII-RNGILSQGPN---------YLPESLRILEWHRHPS 1944
Query: 594 RSFPLDF 600
P DF
Sbjct: 1945HCLPSDF 1965
>TC229101 disease resistance-like protein
Length = 2755
Score = 333 bits (854), Expect = 3e-91
Identities = 264/836 (31%), Positives = 402/836 (47%), Gaps = 35/836 (4%)
Frame = +1
Query: 218 GMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQKIDRYGIYSLRKKYLSKLLGED----- 272
GMGG+GK+TLA A+YH F + C + N ++ ++G+ L++ L+++ E+
Sbjct: 1 GMGGVGKTTLARAVYHSAAGHFDTSCFLANVRENAMKHGLVHLQQTLLAEIFRENNIRLT 180
Query: 273 IQSNGLNYAIERVKRAKVLLILDDLKISIPILELIGGHGNFGQGSRIIVTSRHRHVLKNA 332
G++ +R+ R ++LL+LDD+ + L+G FG GS++I+T+R RH+LK
Sbjct: 181 SVEQGISLIKKRLHRKRLLLVLDDVCELDDLRALVGSPDWFGPGSKVIITTRDRHLLKAH 360
Query: 333 EADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLY 392
D++YEV+ ++ ++LEL AF+ D + N + + +A G+PLAL+++GS LY
Sbjct: 361 GVDKVYEVEVLANGEALELLCWKAFRTDRVHPDFINKLNRAITFASGIPLALELIGSSLY 540
Query: 393 DKEREVWESELKKLKELPNVDIFDVLKLSYDGLDDEPKDIFLDITCFYAGDFVDKVVELL 452
+ E WES L + ++ P DI LK+S+D L K++FLDI CF+ G + ++ +L
Sbjct: 541 GRGIEEWESTLDQYEKNPPRDIHMALKISFDALGYLEKEVFLDIACFFNGFELAEIEHIL 720
Query: 453 DS---CGFSADIGMDVLKDRGLISILE-DRIAVHDLVLEMGREIVRQQCVSDPGKHSHLW 508
+ C IG L ++ LI I E R+ +HDL+ +MGREIVRQ+ PGK S LW
Sbjct: 721 GAHHGCCLKFHIG--ALVEKSLIMIDEHGRVQMHDLIQQMGREIVRQESPEHPGKRSRLW 894
Query: 509 NHKEIYHVVRKNKGTDAIQCIFLDMCQIEK-VQLHPEIFKSMPNIRMLYFHKHSSFKQGQ 567
+ ++I HV+ N GT IQ I LD + EK VQ F M ++R L K F +G
Sbjct: 895 STEDIVHVLEDNTGTCKIQSIILDFSKSEKVVQWDGMAFVKMISLRTLIIRK--MFSKGP 1068
Query: 568 SNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPEKLVKLHMRESRLEQLWGDDQVLQS 627
N + LK+L W P +S P DF PEKL L + S
Sbjct: 1069 KNFQI----------LKMLEWWGCPSKSLPSDFKPEKLAILKLPYSG------------- 1179
Query: 628 LCHGLSYIFFGHLIVFSLHQELPNLERLDLSNSWKLVRIPDLSKSPNIKEIILSGCKSLT 687
F + LH + N +R L R PDLS +P +KE+ C++L
Sbjct: 1180 --------FMSLELPNFLHMRVLNFDRCKF-----LTRTPDLSGAPILKELSSVFCENLV 1320
Query: 688 RLPINL----------------------FKLKFLERLNLSGCSNVENIPEIKETMENLAV 725
+ ++ KL LE +NLS CS++ + PEI MEN+
Sbjct: 1321 EIHDSVGFLDKLEIMNFESCSKLETFPPIKLTSLESINLSHCSSLVSFPEILGKMENITH 1500
Query: 726 LVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYCESLETFP 785
L L+ TAI +LP+S+ LV L+ L L C ++ +PSSI TL +L L + CE L F
Sbjct: 1501 LSLEYTAISKLPNSIRELVRLQSLELHNCGMVQ-LPSSIVTLRELEVLSICQCEGLR-FS 1674
Query: 786 GSIFDLKLTKLDLNDCSRLKTFPA-ILEPAESFAHISLAETA-IKKLPSSLQNLVGLQTL 843
D+K L L S LK ++ F LA A +K L S N L +
Sbjct: 1675 KQDEDVKNKSL-LMPSSYLKQVNLWSCSISDEFIDTGLAWFANVKSLDLSANNFTILPS- 1848
Query: 844 CLKECPDLESLPNSICNLKLLSELD-CSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLP 902
C++EC L L LD C+ ++ GIP ++ LS++R SL+D +
Sbjct: 1849 CIQECRLLRKL-----------YLDYCTHLHEIRGIPPNLETLSAIRCTSLKDLDLAVPL 1995
Query: 903 ESIAHLSSLESLDVSYCRKLECIPHLPPFMKQLLAFHCPSIRSVMSNSRFLSDSKEGTFE 962
ES L L + C L+ I +PP ++ L A +C S+ + S R L E
Sbjct: 1996 ESTKEGCCLRQLILDDCENLQEIRGIPPSIEFLSATNCRSLTA--SCRRMLLKQ-----E 2154
Query: 963 LYFTNNEKQDPGAQRDVVRNAEHRITDVACRFAFYCFPGSEVPNWFHHHCKRNSVT 1018
L+ N++ Y PG+ +P WF H + S++
Sbjct: 2155 LHEAGNKR--------------------------YSLPGTRIPEWFEHCSRGQSIS 2244
>TC221834 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (45%)
Length = 3472
Score = 327 bits (839), Expect = 1e-89
Identities = 203/530 (38%), Positives = 308/530 (57%), Gaps = 22/530 (4%)
Frame = +1
Query: 23 KYDVFLSFSGEDTGKTFTSHLHAALRRKNVFIDDRS-LETGDEIC-------KAIEASTI 74
++DVFLSF GEDT F +L+ AL K R L G+EI KAI+ S +
Sbjct: 85 EFDVFLSFRGEDTRLGFVGNLYKALTEKGFHTFFREKLVRGEEIAASPSVVEKAIQHSRV 264
Query: 75 YVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQ 134
+V+VFS+NYASS CL+EL+ I+ + R V+P+FY VDPS V QT YG+A H+
Sbjct: 265 FVVVFSQNYASSTRCLEELLSILRFSQDNRRPVLPVFYYVDPSDVGLQTGMYGEALAMHE 444
Query: 135 NRL-VPGVVMQRWKKALLEAADLSGY---HSIAARSESMLIDRIAQDIFKKLNPPFSQGM 190
R + +W+KAL EAA LSG+ H E LI++I + + KK+N P
Sbjct: 445 KRFNSESDKVMKWRKALCEAAALSGWPFKHGDGYEYE--LIEKIVEGVSKKINRP----- 603
Query: 191 LGIDKHIAQIQSLLQLDS-EAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQ 249
+G+ + ++ LL S V +IGI G+GGIGK+TLA ALY + QF + C + +
Sbjct: 604 VGLQYRMLELNGLLDAASLSGVHLIGIYGVGGIGKTTLARALYDSVAVQFDALCFLDEVR 783
Query: 250 QKIDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPIL 304
+ ++G+ L++ L++ +GE DI+ G+ +R++ +VLL+LDD+ S +
Sbjct: 784 ENAMKHGLVHLQQTILAETVGEKDIRLPSVKQGITLLKQRLQEKRVLLVLDDINESEQLK 963
Query: 305 ELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLE 364
L+G G FG GSR+I+T+R R +L++ ++IYEV+ ++ ++LEL AFK D
Sbjct: 964 ALVGSPGWFGPGSRVIITTRDRQLLESHGVEKIYEVENLADGEALELLCWKAFKTDKVYP 1143
Query: 365 GYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDG 424
+ N I L YA G+PLAL+V+GS L+ +E W+ L +++ + DI +LK+S+D
Sbjct: 1144DFINKIYRALTYASGLPLALEVIGSNLFGREIVEWQYTLDLYEKIHDKDIQKILKISFDA 1323
Query: 425 LDDEPKDIFLDITCFYAGDFVDKVVELLDS-CGFSADIGMDVLKDRGLISILE-DRIAVH 482
LD+ KD+FLDI CF+ G + +V ++ G S +DVL ++ LI I E R+ +H
Sbjct: 1324LDEHEKDLFLDIACFFKGCKLAQVESIVSGRYGDSLKAIIDVLLEKTLIKIDEHGRVKMH 1503
Query: 483 DLVLEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKN--KGTDAIQCIF 530
DL+ +MGREIVRQ+ PG S LW+ +++ V+ N TD F
Sbjct: 1504DLIQQMGREIVRQESPKHPGNCSRLWSPEDVADVL*GNTVSNTDVFNLTF 1653
Score = 57.8 bits (138), Expect = 3e-08
Identities = 47/172 (27%), Positives = 73/172 (42%), Gaps = 14/172 (8%)
Frame = +1
Query: 648 ELPNLERLDLSNSWK---LVRIPDLSKSPNIKEIILSGCKSLTRLPINLFKLKFLERLNL 704
ELP + + + K L +IPDLS +PN+KE+ C++L + ++ L L+ +N
Sbjct: 2134 ELPKFSHMSVLSFDKFKFLTQIPDLSGTPNLKELSFVLCENLVEIHESVGFLDKLKSMNF 2313
Query: 705 SGCSNVENIPEIKETMENLAVLVLKQTAIQELPSSLNHLVGLEELSLKFCSKLESIPSSI 764
GCS +E P IK L LE ++L +CS L S P +
Sbjct: 2314 EGCSKLETFPPIK-------------------------LTSLESINLSYCSSLVSFPEIL 2418
Query: 765 GTLTK-----------LCKLDLTYCESLETFPGSIFDLKLTKLDLNDCSRLK 805
G + L L L C ++ P SI +L L ++ C L+
Sbjct: 2419 GKMEYTAISILP*LF*LRSLQLHRCGMVQ-LPSSIVMSELEVLSISQCEGLQ 2571
Score = 49.3 bits (116), Expect = 1e-05
Identities = 61/206 (29%), Positives = 84/206 (40%), Gaps = 3/206 (1%)
Frame = +1
Query: 721 ENLAVLVLKQTAIQ--ELPSSLNHLVGLEELSLKFCSKLESIPSSIGTLTKLCKLDLTYC 778
E LA+L L + ELP +H+ L KF L IP GT L +L C
Sbjct: 2086 EKLAILKLPSSCFMSLELPK-FSHMSVLSFDKFKF---LTQIPDLSGT-PNLKELSFVLC 2250
Query: 779 ESLETFPGSIFDL-KLTKLDLNDCSRLKTFPAILEPAESFAHISLAETAIKKLPSSLQNL 837
E+L S+ L KL ++ CS+L+TFP I L
Sbjct: 2251 ENLVEIHESVGFLDKLKSMNFEGCSKLETFPPI-------------------------KL 2355
Query: 838 VGLQTLCLKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTG 897
L+++ L C L S P + G + T I + + L LR+L L G
Sbjct: 2356 TSLESINLSYCSSLVSFPEIL------------GKMEYTAI-SILP*LF*LRSLQLHRCG 2496
Query: 898 VVNLPESIAHLSSLESLDVSYCRKLE 923
+V LP SI +S LE L +S C L+
Sbjct: 2497 MVQLPSSIV-MSELEVLSISQCEGLQ 2571
Score = 40.4 bits (93), Expect = 0.005
Identities = 28/81 (34%), Positives = 36/81 (43%), Gaps = 1/81 (1%)
Frame = +2
Query: 521 KGTDAIQCIFLDMCQIEKV-QLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESL 579
KGT IQ I LD + E+V Q F M N+ L K + + L
Sbjct: 1868 KGTRNIQSIVLDFSKPEEVVQWDGMAFMKMKNLTTLIIRKECFSEDPKK----------L 2017
Query: 580 PNGLKVLHWDEFPQRSFPLDF 600
PN L+VL W +P +S P DF
Sbjct: 2018 PNSLRVLEWRGYPSKSLPSDF 2080
Score = 40.0 bits (92), Expect = 0.006
Identities = 26/84 (30%), Positives = 39/84 (45%)
Frame = +1
Query: 845 LKECPDLESLPNSICNLKLLSELDCSGCRKLTGIPNDIGCLSSLRTLSLQDTGVVNLPES 904
L + PDL PN L EL C L I +G L L++++ + +
Sbjct: 2188 LTQIPDLSGTPN-------LKELSFVLCENLVEIHESVGFLDKLKSMNFEGCSKLETFPP 2346
Query: 905 IAHLSSLESLDVSYCRKLECIPHL 928
I L+SLES+++SYC L P +
Sbjct: 2347 IK-LTSLESINLSYCSSLVSFPEI 2415
>TC229789 homologue to UP|Q9FVK4 (Q9FVK4) Resistance protein LM17 (Fragment),
partial (94%)
Length = 2104
Score = 288 bits (737), Expect = 1e-77
Identities = 205/619 (33%), Positives = 329/619 (53%), Gaps = 27/619 (4%)
Frame = +2
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFLSF G DT FT +L+ AL + FID+ L+ G+EI KAIE S I +I
Sbjct: 146 YDVFLSFRGSDTLHGFTGYLYEALHDSGIHTFIDE-DLKRGEEITPAIVKAIEESRIAII 322
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V S NYASS CLDEL I++C V+P+FY VD V SY +A +H L
Sbjct: 323 VLSINYASSSSCLDELATILDCLKRKRLLVLPVFYNVDHYQVLG--GSYVEALVKHGKSL 496
Query: 138 VPGVV-MQRWKKALLEAADLSGYH-SIAARSESMLIDRIAQDIFKKLNPPFSQGMLGIDK 195
+ +++W+ AL E ADLS + AR E I I + + K+NP + +G+
Sbjct: 497 KHSMEKLEKWEMALYEVADLSDFKIKHGARYEYDFIGEIVEWVSSKINP--AHYPVGLGS 670
Query: 196 HIAQIQSLLQLD-SEAVRIIGICGMGGIGKSTLAEALYHKL-GTQFSSRCLIVNAQQKID 253
+ +++ LL + + V ++GI G+ G+GKSTLA +Y+KL F + C I N ++K
Sbjct: 671 KVLEVRKLLDVGRDDGVHMLGIHGIDGVGKSTLAREVYNKLISDHFDASCFIENVREKSK 850
Query: 254 RYGIYSLRKKYLSKLLGE-DIQSNGLNYAIE-----RVKRAKVLLILDDLKISIPILELI 307
++G++ L+ LSK+LGE DI I R+++ KVL++LDD+ +
Sbjct: 851 KHGLHHLQNILLSKILGEKDINLTSAQQEISMMQRHRLQQKKVLMVLDDVDRPEQLQAGT 1030
Query: 308 GGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGYA 367
G FG GS++I+T++ + +L + + + YEV+ ++ D+L+L + AFK Y Y
Sbjct: 1031GKPAWFGPGSKVIITTQDKQLLTSYDINRTYEVKKLNKDDALQLLKWKAFKMHYFDPRYK 1210
Query: 368 NLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLDD 427
L+ + YA +PL +++L S L+ K + W+S + + PN I +LK+ +D L +
Sbjct: 1211KLLNHAVTYASSLPLTMEILASNLFGKSVKEWKSTFHQFERSPNNPIEMILKVIFDSLKE 1390
Query: 428 EPKDIFLDITCFYAGDFVDKVVELLDS-CGFSADIGMDVLKDRGLISI------LEDRIA 480
+ K + LDI C++ G + +V ++L + G +DVL D+ L+ I D I
Sbjct: 1391KEKSVLLDIACYFKGYELTEVQDILHAHYGQCMKYYIDVLVDKSLVYITHGTEPCNDTIT 1570
Query: 481 VHDLVLEMGREIVR-QQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLD---MCQI 536
+H+L+ +EIVR + ++ PG+ LW+ +++ V K I+ I LD +
Sbjct: 1571MHELI---AKEIVRLESMMTKPGECRRLWSWEDVREVFSK------IEIICLDYPIFDEE 1723
Query: 537 EKVQLHPEIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSF 596
E VQ F++M N++ L ++ +F +G E LPN L+V W +P
Sbjct: 1724EIVQWDGTAFQNMQNLKTLII-RNGNFSKGP---------EYLPNSLRVFEWWGYPSHCL 1873
Query: 597 PLDFCPEKLVKLHMRESRL 615
P DF P++L + SR+
Sbjct: 1874PSDFHPKELAICKLPCSRI 1930
>NP595166 R 14 protein [Glycine max]
Length = 1926
Score = 273 bits (698), Expect = 3e-73
Identities = 165/425 (38%), Positives = 247/425 (57%), Gaps = 17/425 (4%)
Frame = +1
Query: 21 PLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTI 74
P YDVFLSF GEDT FT +L+ AL K + F D+ L GDEI KAI+ S I
Sbjct: 25 PFIYDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSKAIQESRI 204
Query: 75 YVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQ 134
+ V S+NYA S CLDELV I+ CK VIP+FY VDPS +RHQ SYG+A +HQ
Sbjct: 205 AITVLSQNYAFSSFCLDELVTILHCKSE-GLLVIPVFYNVDPSDLRHQKGSYGEAMIKHQ 381
Query: 135 NRLVPGVV-MQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPPFSQGM-- 190
R + +Q+W+ AL + ADLSG+H + E I I +++ +K+N +
Sbjct: 382 KRFESKMEKLQKWRMALKQVADLSGHHFKDGDAYEYKFIGSIVEEVSRKINRASLHVLDY 561
Query: 191 -LGIDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNA 248
+G++ + + LL + S+ V IIGI GM G+GK+TL+ A+Y+ + F C + N
Sbjct: 562 PVGLESQVTDLMKLLDVGSDDVVHIIGIHGMRGLGKTTLSLAVYNLIALHFDESCFLQNV 741
Query: 249 QQKIDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPI 303
+++ +++G+ L+ L KLLGE DI G + R++R KVLLILDD +
Sbjct: 742 REESNKHGLKHLQSILLLKLLGEKDINLTSWQEGASMIQHRLRRKKVLLILDDADRHEQL 921
Query: 304 LELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPL 363
++G FG GSR+I+T+R +H+LK + YEV+ ++ +L+L NAF+ +
Sbjct: 922 KAIVGRPDWFGPGSRVIITTRDKHLLKYHGIERTYEVKVLNDNAALQLLTWNAFRREKID 1101
Query: 364 EGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYD 423
Y +++ V+ YA G+PLAL+V+GS L++K WE ++ +P +I D+LK+S+D
Sbjct: 1102PSYEHVLNRVVAYASGLPLALEVIGSHLFEKTVAEWEYAVEHYSRIPIDEIVDILKVSFD 1281
Query: 424 GLDDE 428
E
Sbjct: 1282ATKQE 1296
>NP595167 R 13 protein [Glycine max]
Length = 1926
Score = 263 bits (673), Expect = 3e-70
Identities = 161/417 (38%), Positives = 241/417 (57%), Gaps = 17/417 (4%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFLSF G DT FT +L+ AL + + FIDD+ L GD+I AI S I +
Sbjct: 34 YDVFLSFRGLDTRNGFTGNLYKALGDRGIYTFIDDQELPRGDKITPALSNAINESRIAIT 213
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V SENYA S CLDELV I+ CK VIP+FY+VDPS VRHQ SYG+ +HQ R
Sbjct: 214 VLSENYAFSSFCLDELVTILHCKSE-GLLVIPVFYKVDPSDVRHQKGSYGETMTKHQKRF 390
Query: 138 VPGVV-MQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPP---FSQGMLG 192
+ ++ W+ AL + ADLSGYH S E I I +++ +K+N + +
Sbjct: 391 ESKMEKLREWRMALQQVADLSGYHFKDGDSYEYKFIGNIVEEVSRKINHASLHVADYPVD 570
Query: 193 IDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQK 251
++ + +++ LL + S+ V IIGI GM G+GK+TLA A+Y+ + F C + N +++
Sbjct: 571 LESQVIEVRKLLDVGSDDVVHIIGIHGMRGLGKTTLALAVYNLIALHFDESCFLQNVREE 750
Query: 252 IDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
+++G+ L+ L KLLGE DI +G + R+++ KVLLILDD + +
Sbjct: 751 SNKHGLKHLQSILLLKLLGEKDITLTSWQDGASMIQRRLRQKKVLLILDDADEQEQLKAI 930
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
+G FG GSR+I+T+R +H+LK + YEV+ ++ +L+L NAFK + Y
Sbjct: 931 VGSPNCFGPGSRVIITTRDKHLLKYHGVERTYEVKVLNQNAALQLLTWNAFKSEKIDPCY 1110
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYD 423
+++ V+ YA G+P AL+ +GS L+ K WE ++ K +P +I + KLS+D
Sbjct: 1111EDVLNRVVAYASGLPRALEAIGSNLFGKTVAEWEYAVEHYKTIPRDEILESPKLSFD 1281
>TC221449 UP|Q9FXZ1 (Q9FXZ1) Resistance protein MG23 (Fragment), complete
Length = 1307
Score = 255 bits (651), Expect = 9e-68
Identities = 154/428 (35%), Positives = 246/428 (56%), Gaps = 21/428 (4%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEICKA----IEASTIYVI 77
YDVFLSF GEDT FT +L+ LR + + FIDD L+ G EI KA IE S I++I
Sbjct: 22 YDVFLSFRGEDTRHGFTGNLYNVLRERGIDTFIDDEELQKGHEITKALEEAIEKSKIFII 201
Query: 78 VFSENYASSRCCLDELVKIME-CKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNR 136
V SENYASS CL+EL I+ K DR+++P+FY+VDPS VR+ S+G+A H+ +
Sbjct: 202 VLSENYASSSFCLNELTHILNFTKGKSDRSILPVFYKVDPSDVRYHRGSFGEALANHEKK 381
Query: 137 LVPGVV--MQRWKKALLEAADLSGYH--SIAARSESMLIDRIAQDIFKKLNPPF---SQG 189
L + +Q WK AL + ++ SG+H + E I I + + K N S
Sbjct: 382 LKSNYMEKLQIWKMALQQVSNFSGHHFQPDGDKYEYDFIKEIVESVPSKFNRNLLYVSDV 561
Query: 190 MLGIDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNA 248
++G+ + ++SLL + ++ V ++GI G+GG+GK+TLA A+Y+ + F + C + N
Sbjct: 562 LVGLKSPVLAVKSLLDVGADDVVHMVGIHGLGGVGKTTLAVAVYNSIACHFEACCFLENV 741
Query: 249 QQKIDRYGIYSLRKKYLSKLLGE-----DIQSNGLNYAIERVKRAKVLLILDDLKISIPI 303
++ ++ G+ SL+ LSK +G+ G + ++K KVLL+LDD+ +
Sbjct: 742 RETSNKKGLESLQNILLSKTVGDMKIEVTNSREGTDIIKRKLKEKKVLLVLDDVNEHEQL 921
Query: 304 LELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPL 363
+I FG+GSR+I+T+R +L Y+V+ ++ + +L+L AF + +
Sbjct: 922 QAIIDSPDWFGRGSRVIITTRDEQLLVLHNVKRTYKVRELNEKHALQLLTQKAFGLEKKV 1101
Query: 364 E-GYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSY 422
+ Y +++ + YA G+PLAL+V+GS L+ K E WES L + P+ I+ LK+SY
Sbjct: 1102DPSYHDILNRAVTYASGLPLALKVIGSNLFGKSIEEWESVLDGYERSPDKSIYMTLKVSY 1281
Query: 423 DGLDDEPK 430
D L+++ K
Sbjct: 1282DALNEDEK 1305
>TC231313 UP|Q9FXZ2 (Q9FXZ2) Resistance protein LM12 (Fragment), complete
Length = 1314
Score = 251 bits (642), Expect = 1e-66
Identities = 155/431 (35%), Positives = 247/431 (56%), Gaps = 20/431 (4%)
Frame = +1
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGDEICKAIEA----STIYVI 77
YDVFLSF GEDT FT +L+ LR + + FIDD + GDEI A+EA S I++I
Sbjct: 22 YDVFLSFRGEDTRYGFTGYLYNVLRERGIHTFIDDDEPQEGDEITTALEAAIEKSKIFII 201
Query: 78 VFSENYASSRCCLDELVKIME-CKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNR 136
V SENYASS CL+ L I+ K N D V+P+FYRV+PS VRH S+G+A H+ +
Sbjct: 202 VLSENYASSSFCLNSLTHILNFTKENNDVLVLPVFYRVNPSDVRHHRGSFGEALANHEKK 381
Query: 137 LVPGVV--MQRWKKALLEAADLSGYH--SIAARSESMLIDRIAQDIFKKLNPPF---SQG 189
+ ++ WK AL + +++SG+H + E I I + + K N S
Sbjct: 382 SNSNNMEKLETWKMALHQVSNISGHHFQHDGNKYEYKFIKEIVESVSNKFNHDHLHVSDV 561
Query: 190 MLGIDKHIAQIQSLLQLD-SEAVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNA 248
++G++ + +++SLL + + V ++GI G+ G+GK+TLA A+Y+ + F + C + N
Sbjct: 562 LVGLESPVLEVKSLLDVGRDDVVHMVGIHGLAGVGKTTLAIAVYNSIAGHFEASCFLENV 741
Query: 249 QQKIDRY-GIYSLRKKYLSKLLGEDIQSN---GLNYAIERVKRAKVLLILDDLKISIPIL 304
++ + G+ L+ LSK GE +N G+ ++K+ K+LLILDD+ +
Sbjct: 742 KRTSNTINGLEKLQSFLLSKTAGEIKLTNWREGIPIIKRKLKQKKILLILDDVDEDKQLQ 921
Query: 305 ELIGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLE 364
LIG FG GSRII+T+R H+L Y+V+ ++ + +L+L AF+ + ++
Sbjct: 922 ALIGSPDWFGLGSRIIITTRDEHLLALHNVKITYKVRELNEKHALQLLTQKAFELEKGID 1101
Query: 365 -GYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYD 423
Y +++ + YA G+P L+V+GS L+ K E W+S L + +P+ +LK+SYD
Sbjct: 1102PSYHDILNRAVTYASGLPFVLEVIGSNLFGKSIEEWKSALDGYERIPHKKNLCILKVSYD 1281
Query: 424 GLDDEPKDIFL 434
L+++ K IFL
Sbjct: 1282ALNEDEKSIFL 1314
>TC227729
Length = 1391
Score = 249 bits (635), Expect = 6e-66
Identities = 155/425 (36%), Positives = 237/425 (55%), Gaps = 23/425 (5%)
Frame = +2
Query: 6 SSSSAAAAAMAPPPNPLKYDVFLSFSGEDTGKTFTSHLHAALRRKNV--FIDDRSLETGD 63
+S+S A + +YDVF+SF GEDT +FT L AL+++ + F DD+ + G+
Sbjct: 119 ASTSNAIIQCTSSSSSFEYDVFVSFRGEDTRNSFTGFLFEALKKQGIEAFKDDKDIRKGE 298
Query: 64 ----EICKAIEASTIYVIVFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTV 119
E+ +AIE S ++++VFS++YASS CL EL I C R ++PIFY VDPS V
Sbjct: 299 SIAPELIRAIEGSHVFLVVFSKDYASSTWCLRELAHIWNCIQTSSRLLLPIFYDVDPSQV 478
Query: 120 RHQTNSYGDAFDEH-QNRLVPGVVMQRWKKALLEAADLSGYHSIAARSESMLIDRIAQDI 178
R Q+ Y AF +H Q+ ++ W++ L + A LSG+ I + + +I+ I Q I
Sbjct: 479 RKQSGDYEKAFSQHQQSSRFQEKEIKTWREVLKQVASLSGW-DIRNKQQHPVIEEIVQQI 655
Query: 179 FKKLNPPFS----QGMLGIDKHIAQIQSLLQLD--SEAVRIIGICGMGGIGKSTLAEALY 232
L FS ++G++ H A++ L+ ++ VR++GI GMGGIGKSTL ALY
Sbjct: 656 KNILGCKFSILPYDYLVGMESHFAKLSKLICPGPVNDDVRVVGITGMGGIGKSTLGRALY 835
Query: 233 HKLGTQFSSRCLIVNAQQKIDRYGIYSLRKKYLSKLLGED-----IQSNGLNYAIERVKR 287
++ QF+SRC I + + YG ++K+ LS+ L E SNG ER+
Sbjct: 836 ERISHQFNSRCYIDDVSKLYQGYGTLGVQKELLSQSLNEKNLKICNVSNGTLLVWERLSN 1015
Query: 288 AKVLLILDDLKISIPILELIGGHGN-----FGQGSRIIVTSRHRHVLKNAEADEIYEVQG 342
AK L+ILD++ + GG + G+GS +I+ SR + +LK D IY V+
Sbjct: 1016AKALIILDNVDQDKQLDMFTGGRNDLLGKCLGKGSIVIIISRDQQILKAHGVDVIYRVEP 1195
Query: 343 MSYQDSLELFRLNAFKEDYPLEGYANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESE 402
++ D+L LF AFK +Y + + L +VL + +G PLA++VLGS L+ K+ W S
Sbjct: 1196LNDNDALGLFCKKAFKNNYMMSDFKKLTSDVLSHCQGHPLAIEVLGSSLFGKDVSHWGSA 1375
Query: 403 LKKLK 407
L L+
Sbjct: 1376LVSLR 1390
>TC224280 UP|Q9FVK3 (Q9FVK3) Resistance protein MG63 (Fragment), complete
Length = 1379
Score = 241 bits (614), Expect = 2e-63
Identities = 143/422 (33%), Positives = 241/422 (56%), Gaps = 7/422 (1%)
Frame = +1
Query: 191 LGIDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQ 249
+G++ I +++ LL + S+ V ++GI G+GGIGK+TLA A+Y+ + F + C + N +
Sbjct: 46 VGLESRIQEVKMLLDVGSDDVVHMVGIHGLGGIGKTTLAAAIYNSIADHFEALCFLENVR 225
Query: 250 QKIDRYGIYSLRKKYLSKLLGEDIQ---SNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
+ +G+ L++ LS+ +GED G++ R+++ KVLLILDD+ + L
Sbjct: 226 ETSKTHGLQYLQRNLLSETVGEDELIGVKQGISIIQHRLQQKKVLLILDDVDKREQLQAL 405
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMSYQDSLELFRLNAFKEDYPLEGY 366
+G F GSR+I+T+R + +L YEV ++ + +L+L AFK + Y
Sbjct: 406 VGRPDLFCPGSRVIITTRDKQLLACHGVKRTYEVNELNEEYALQLLSWKAFKLEKVNPCY 585
Query: 367 ANLIVEVLKYAKGVPLALQVLGSLLYDKEREVWESELKKLKELPNVDIFDVLKLSYDGLD 426
+++ + Y+ G+PLAL+V+GS L + E W S L + K +PN +I ++LK+SYD L+
Sbjct: 586 KDVLNRTVTYSAGLPLALEVIGSNLSGRNIEQWRSTLDRYKRIPNKEIQEILKVSYDALE 765
Query: 427 DEPKDIFLDITCFYAGDFVDKVVELLDS-CGFSADIGMDVLKDRGLISILEDRIAVHDLV 485
++ + +FLDI+C + +V ++L + G + + VL ++ LI I + I +HDL+
Sbjct: 766 EDEQSVFLDISCCLKEYDLKEVQDILRAHYGHCMEHHIRVLLEKSLIKISDGYITLHDLI 945
Query: 486 LEMGREIVRQQCVSDPGKHSHLWNHKEIYHVVRKNKGTDAIQCIFLDMCQIEKVQLH--P 543
+MG+EIVR++ +PGK S LW H +I +GT I+ I D E+V++
Sbjct: 946 EDMGKEIVRKESPREPGKRSRLWLHTDII------QGTSQIEIICTDFSLFEEVEIEWDA 1107
Query: 544 EIFKSMPNIRMLYFHKHSSFKQGQSNVIVPDLLESLPNGLKVLHWDEFPQRSFPLDFCPE 603
FK M N++ L K+ F +G + LP+ L+VL W +P +SFP DF P+
Sbjct: 1108NAFKKMENLKTLII-KNGHFTKGPKH---------LPDTLRVLEWWRYPSQSFPSDFRPK 1257
Query: 604 KL 605
KL
Sbjct: 1258KL 1263
>TC209219 UP|Q9FVK2 (Q9FVK2) Resistance protein MG13 (Fragment), complete
Length = 1224
Score = 214 bits (544), Expect = 2e-55
Identities = 135/338 (39%), Positives = 196/338 (57%), Gaps = 17/338 (5%)
Frame = +3
Query: 24 YDVFLSFSGEDTGKTFTSHLHAALRRK--NVFIDDRSLETGDEIC----KAIEASTIYVI 77
YDVFLSF G DT FT +L+ AL K + F D+ L +G+EI KAI+ S + +I
Sbjct: 63 YDVFLSFRGTDTRYGFTGNLYKALCDKGFHTFFDEDKLHSGEEITPALLKAIQDSRVAII 242
Query: 78 VFSENYASSRCCLDELVKIMECKMNYDRAVIPIFYRVDPSTVRHQTNSYGDAFDEHQNRL 137
V SENYA S CLDELV I CK VIP+FY+VDPS VRHQ SYG+A +HQ R
Sbjct: 243 VLSENYAFSSFCLDELVTIFHCKRE-GLLVIPVFYKVDPSYVRHQKGSYGEAMTKHQERF 419
Query: 138 VPGVV-MQRWKKALLEAADLSGYHSIAARS-ESMLIDRIAQDIFKKLNPP---FSQGMLG 192
+ +Q W+ AL + ADLSG H S E I I +++ +K+ + +G
Sbjct: 420 KDKMEKLQEWRMALKQVADLSGSHFKDGGSYEYEFIGSIVEEVSRKIGRGSLHVADYPVG 599
Query: 193 IDKHIAQIQSLLQLDSE-AVRIIGICGMGGIGKSTLAEALYHKLGTQFSSRCLIVNAQQK 251
+ ++ LL + S+ V IIGI GM G+GK+TLA +Y+ + F C + N +++
Sbjct: 600 QASQVTEVMKLLDVGSDDVVHIIGIYGMRGLGKTTLALDVYNSIARHFDESCFLQNVREE 779
Query: 252 IDRYGIYSLRKKYLSKLLGE-DIQ----SNGLNYAIERVKRAKVLLILDDLKISIPILEL 306
++G+ L+ +SKLLGE DI G + R++R KVLLILDD+ + +
Sbjct: 780 SKKHGLKHLQSIIISKLLGEKDINLASYREGASMIQSRLRRKKVLLILDDVNKREQLKAI 959
Query: 307 IGGHGNFGQGSRIIVTSRHRHVLKNAEADEIYEVQGMS 344
+G FG GSR+I+T+R++ +LK+ E + Y+V+ +S
Sbjct: 960 VGRSDWFGPGSRVIITTRYKRLLKDHEVERTYKVKLLS 1073
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,376,775
Number of Sequences: 63676
Number of extensions: 848412
Number of successful extensions: 5769
Number of sequences better than 10.0: 308
Number of HSP's better than 10.0 without gapping: 4782
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5274
length of query: 1156
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1048
effective length of database: 5,762,624
effective search space: 6039229952
effective search space used: 6039229952
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0073.8


