

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.3
(191 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
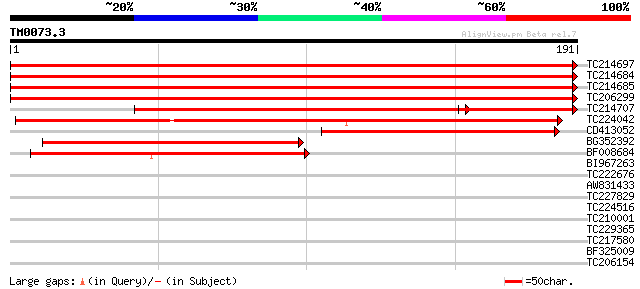
Score E
Sequences producing significant alignments: (bits) Value
TC214697 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein ... 359 e-100
TC214684 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein ... 359 e-100
TC214685 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein ... 358 e-100
TC206299 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein ... 344 1e-95
TC214707 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein ... 202 8e-53
TC224042 weakly similar to UP|Q8LD03 (Q8LD03) 40S ribosomal prot... 187 2e-48
CD413052 similar to SP|Q9ZNS1|RS7_A 40S ribosomal protein S7. [G... 130 5e-31
BG352392 similar to SP|Q9ZNS1|RS7_A 40S ribosomal protein S7. [G... 114 4e-26
BF008684 84 5e-17
BI967263 similar to GP|27446644|gb| mu opioid receptor variant M... 27 0.047
TC222676 UP|Q84L88 (Q84L88) Apyrase, complete 29 1.2
AW831433 similar to GP|14787424|emb Ribosomal protein S7 {Hordeu... 28 3.4
TC227829 UP|CAP2_SOYBN (P51061) Phosphoenolpyruvate carboxylase ... 27 4.4
TC224516 27 4.4
TC210001 similar to UP|Q9MA74 (Q9MA74) Histidine decarboxylase (... 27 5.8
TC229365 27 7.5
TC217580 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (74%) 26 9.8
BF325009 26 9.8
TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division prote... 26 9.8
>TC214697 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein S7, complete
Length = 934
Score = 359 bits (922), Expect = e-100
Identities = 178/191 (93%), Positives = 187/191 (97%)
Frame = +2
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
MYTSRKKIHKDKDAEPTEFEE+VGQALFDLENT+ ELKSDLKDLYINSAVQ+DVSGNRKA
Sbjct: 74 MYTSRKKIHKDKDAEPTEFEESVGQALFDLENTNNELKSDLKDLYINSAVQIDVSGNRKA 253
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVIHVPYRLRK FRK+HV+LVRELEKKFSGKDVILIATRRILRPPKKGSA QRPRTRTLT
Sbjct: 254 VVIHVPYRLRKGFRKIHVRLVRELEKKFSGKDVILIATRRILRPPKKGSAVQRPRTRTLT 433
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
AVHEAMLED+VLPAEIVGKRVRY++DGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK
Sbjct: 434 AVHEAMLEDIVLPAEIVGKRVRYKIDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 613
Query: 181 DVVFEYPITEA 191
D+VFEYP TEA
Sbjct: 614 DIVFEYPTTEA 646
>TC214684 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein S7, partial
(98%)
Length = 836
Score = 359 bits (922), Expect = e-100
Identities = 178/191 (93%), Positives = 190/191 (99%)
Frame = +2
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
MYTSRKKIHKDKDAEPTEFEE+VGQALFDLENT+QELKSDLKDLYINSAVQVDVSGNRKA
Sbjct: 98 MYTSRKKIHKDKDAEPTEFEESVGQALFDLENTNQELKSDLKDLYINSAVQVDVSGNRKA 277
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVIHVPYRLRK FRK+HV+LVRELEKKFSGKDVIL+ATRRILRPPKKGSAAQRPR+RTLT
Sbjct: 278 VVIHVPYRLRKGFRKIHVRLVRELEKKFSGKDVILVATRRILRPPKKGSAAQRPRSRTLT 457
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
AVHEAMLEDVVLPAEIVGKRVRY++DGSKIMKV+LDPKERNNTEYKLETFAAVYRKLSGK
Sbjct: 458 AVHEAMLEDVVLPAEIVGKRVRYQIDGSKIMKVYLDPKERNNTEYKLETFAAVYRKLSGK 637
Query: 181 DVVFEYPITEA 191
DVVFEYP++E+
Sbjct: 638 DVVFEYPVSES 670
>TC214685 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein S7, complete
Length = 857
Score = 358 bits (919), Expect = e-100
Identities = 177/191 (92%), Positives = 187/191 (97%)
Frame = +1
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
MYTSRKKIHKDKDAEPTEFEE+VGQALFDLENT+ ELKSDLKDLYINSAVQ+DVSGNRKA
Sbjct: 76 MYTSRKKIHKDKDAEPTEFEESVGQALFDLENTNNELKSDLKDLYINSAVQIDVSGNRKA 255
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
V+IHVPYRLRK FRK+HV+LVRELEKKFSGKDVILIATRRI+RPPKKGSA QRPRTRTLT
Sbjct: 256 VIIHVPYRLRKGFRKIHVRLVRELEKKFSGKDVILIATRRIVRPPKKGSAVQRPRTRTLT 435
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
AVHEAMLED+VLPAEIVGKRVRY++DGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK
Sbjct: 436 AVHEAMLEDIVLPAEIVGKRVRYKIDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 615
Query: 181 DVVFEYPITEA 191
DVVFEYP TEA
Sbjct: 616 DVVFEYPTTEA 648
>TC206299 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein S7, partial
(98%)
Length = 1036
Score = 344 bits (883), Expect = 1e-95
Identities = 168/191 (87%), Positives = 185/191 (95%)
Frame = +2
Query: 1 MYTSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKA 60
MYTSRKKIHKDKDAEPTEFEETVGQ LFDLENT+Q+LKSDLKDLYIN A+Q+DV+GNRKA
Sbjct: 170 MYTSRKKIHKDKDAEPTEFEETVGQYLFDLENTNQDLKSDLKDLYINQAIQMDVAGNRKA 349
Query: 61 VVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLT 120
VVI+VP+RLRKAFRK+H++LVRELEKKFSGKDV+LIATRRI+RPPKKGSA QRPRTRTLT
Sbjct: 350 VVIYVPFRLRKAFRKIHLRLVRELEKKFSGKDVVLIATRRIVRPPKKGSAVQRPRTRTLT 529
Query: 121 AVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSGK 180
AVH+AMLEDVV PAEIVGKR RYR+DGSKIMKVFLDPKERNNTEYKLETF+ VYRKL+GK
Sbjct: 530 AVHDAMLEDVVYPAEIVGKRARYRIDGSKIMKVFLDPKERNNTEYKLETFSGVYRKLTGK 709
Query: 181 DVVFEYPITEA 191
DVVFEYPI+EA
Sbjct: 710 DVVFEYPISEA 742
>TC214707 similar to UP|RS7_AVIMR (Q9ZNS1) 40S ribosomal protein S7, partial
(76%)
Length = 749
Score = 202 bits (514), Expect = 8e-53
Identities = 102/113 (90%), Positives = 108/113 (95%)
Frame = +3
Query: 43 DLYINSAVQVDVSGNRKAVVIHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRIL 102
DLYI AVQVDVSGNRKA VIHVPYRLRK FRK+HV+LVRELEKKFSGKDVIL+ATRRIL
Sbjct: 3 DLYIXXAVQVDVSGNRKAXVIHVPYRLRKGFRKIHVRLVRELEKKFSGKDVILVATRRIL 182
Query: 103 RPPKKGSAAQRPRTRTLTAVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFL 155
RPPKKGSAAQRPR+RTLTAVHEAMLEDVVLPAEIVGKRVRY++DGSKIMKV L
Sbjct: 183 RPPKKGSAAQRPRSRTLTAVHEAMLEDVVLPAEIVGKRVRYQIDGSKIMKVEL 341
Score = 78.2 bits (191), Expect = 2e-15
Identities = 35/40 (87%), Positives = 40/40 (99%)
Frame = +2
Query: 152 KVFLDPKERNNTEYKLETFAAVYRKLSGKDVVFEYPITEA 191
+V+LDPKERNNTEYKLETFAAVYRKLSGKDVVFEYP++E+
Sbjct: 464 QVYLDPKERNNTEYKLETFAAVYRKLSGKDVVFEYPVSES 583
>TC224042 weakly similar to UP|Q8LD03 (Q8LD03) 40S ribosomal protein S7-like,
partial (76%)
Length = 610
Score = 187 bits (476), Expect = 2e-48
Identities = 97/187 (51%), Positives = 137/187 (72%), Gaps = 3/187 (1%)
Frame = +1
Query: 3 TSRKKIHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAVV 62
++R+KI K+K P FE+ V L+DLE + +LKS+L+ L I+SA ++DV GNRKAV+
Sbjct: 19 SARRKIVKEKGQVPDAFEDQVATYLYDLEQSSSDLKSELRHLQISSAKEIDV-GNRKAVI 195
Query: 63 IHVPYRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRILRPPKKGSAAQ---RPRTRTL 119
+ VP+RL F +V +LV ELEKK SGK V+++A RRIL+ + + + RP +RTL
Sbjct: 196 LVVPFRLLNNFHRVQPRLVHELEKKLSGKQVVVVAQRRILQKVGRNNRVKQQKRPVSRTL 375
Query: 120 TAVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEYKLETFAAVYRKLSG 179
TAVH+A+LED V P EIVGKR+R+++DGSK++KV+LD K++ E KLETFA VY+KL+G
Sbjct: 376 TAVHDAILEDTVFPCEIVGKRLRFKLDGSKLIKVYLDKKDQQTYEDKLETFATVYKKLTG 555
Query: 180 KDVVFEY 186
K VF +
Sbjct: 556 KSAVFTF 576
>CD413052 similar to SP|Q9ZNS1|RS7_A 40S ribosomal protein S7. [Grey
mangrove] {Avicennia marina}, partial (36%)
Length = 625
Score = 130 bits (326), Expect = 5e-31
Identities = 64/80 (80%), Positives = 69/80 (86%)
Frame = -3
Query: 106 KKGSAAQRPRTRTLTAVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNNTEY 165
KK +RTLTAVH+AMLEDVV PAEIVGKR RYR+DGSKIMKVFLDPKERNNTEY
Sbjct: 398 KKKKKKTETSSRTLTAVHDAMLEDVVYPAEIVGKRARYRIDGSKIMKVFLDPKERNNTEY 219
Query: 166 KLETFAAVYRKLSGKDVVFE 185
KLETF+ VYRKL+GKDVVFE
Sbjct: 218 KLETFSGVYRKLTGKDVVFE 159
>BG352392 similar to SP|Q9ZNS1|RS7_A 40S ribosomal protein S7. [Grey
mangrove] {Avicennia marina}, partial (31%)
Length = 265
Score = 114 bits (284), Expect = 4e-26
Identities = 59/88 (67%), Positives = 69/88 (78%)
Frame = +2
Query: 12 KDAEPTEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVPYRLRK 71
KDAEPTEFEE+ QAL DLENT+ +LKSDLKDLY+NSA Q+DVSGN +AV IH+PYRLRK
Sbjct: 2 KDAEPTEFEESDRQALPDLENTNNDLKSDLKDLYMNSAAQIDVSGNHEAVGIHMPYRLRK 181
Query: 72 AFRKVHVKLVRELEKKFSGKDVILIATR 99
K HV L ++ GKDV+LIATR
Sbjct: 182RLAKNHVMLCETT*QEGYGKDVVLIATR 265
>BF008684
Length = 285
Score = 83.6 bits (205), Expect = 5e-17
Identities = 51/95 (53%), Positives = 63/95 (65%), Gaps = 1/95 (1%)
Frame = +1
Query: 8 IHKDKDAEPTEFEETVGQALFDLENTHQELKSDLKDLYI-NSAVQVDVSGNRKAVVIHVP 66
IHKD DAEP+EFEE VGQA DLEN++ EL + +Y ++ G RKAV+IHV
Sbjct: 1 IHKD*DAEPSEFEEPVGQA*SDLENSNNELI*RSE*IYT*TQLCKLMFIGIRKAVIIHVR 180
Query: 67 YRLRKAFRKVHVKLVRELEKKFSGKDVILIATRRI 101
YRL FRK+HVKLVR GK+VILI+ RR+
Sbjct: 181 YRLWIRFRKIHVKLVRNPVYIV*GKNVILISARRL 285
>BI967263 similar to GP|27446644|gb| mu opioid receptor variant MOR-1R {Mus
musculus}, partial (3%)
Length = 367
Score = 27.3 bits (59), Expect(2) = 0.047
Identities = 15/29 (51%), Positives = 20/29 (68%)
Frame = +3
Query: 32 NTHQELKSDLKDLYINSAVQVDVSGNRKA 60
+TH L SDL+DLYI A +D +GN +A
Sbjct: 228 STH--LISDLRDLYIIHAXXMDEAGNSQA 308
Score = 25.4 bits (54), Expect(2) = 0.047
Identities = 9/19 (47%), Positives = 15/19 (78%)
Frame = +1
Query: 59 KAVVIHVPYRLRKAFRKVH 77
+ V+++ P +LRKAFR +H
Sbjct: 304 RPVLLYGPLKLRKAFRTIH 360
>TC222676 UP|Q84L88 (Q84L88) Apyrase, complete
Length = 1368
Score = 29.3 bits (64), Expect = 1.2
Identities = 14/48 (29%), Positives = 27/48 (56%)
Frame = -1
Query: 32 NTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVPYRLRKAFRKVHVK 79
N H +++ L + + V ++V G +V H+PYRL+ FR + ++
Sbjct: 528 NCHP*IRTFLGSINYRNCVWLNVEGT--SVAQHIPYRLQYLFRSISIQ 391
>AW831433 similar to GP|14787424|emb Ribosomal protein S7 {Hordeum vulgare
subsp. vulgare}, partial (7%)
Length = 241
Score = 27.7 bits (60), Expect = 3.4
Identities = 11/15 (73%), Positives = 14/15 (93%)
Frame = +2
Query: 47 NSAVQVDVSGNRKAV 61
N A+Q+DV+GNRKAV
Sbjct: 146 NQAIQMDVAGNRKAV 190
>TC227829 UP|CAP2_SOYBN (P51061) Phosphoenolpyruvate carboxylase (PEPCase) ,
complete
Length = 3192
Score = 27.3 bits (59), Expect = 4.4
Identities = 19/94 (20%), Positives = 45/94 (47%), Gaps = 5/94 (5%)
Frame = +1
Query: 17 TEFEETVGQALFDLENTHQELKSDLKDLYINSAVQVDVSGNRKAVVIHVPYRLRKAFRKV 76
++ EET+ + +FDL+ + QE+ LK+ ++ + + + + ++ R+R ++
Sbjct: 448 SDIEETLKKLVFDLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRSLLQKHGRIRNCLSQL 627
Query: 77 HVKLV-----RELEKKFSGKDVILIATRRILRPP 105
+ K + +EL++ + T I R P
Sbjct: 628 YAKDITPDDKQELDEALQREIQAAFRTDEIRRTP 729
>TC224516
Length = 464
Score = 27.3 bits (59), Expect = 4.4
Identities = 21/79 (26%), Positives = 36/79 (44%), Gaps = 1/79 (1%)
Frame = +2
Query: 103 RPPKKGSAAQRPRTRTLTAVHEAMLEDVVLPAEIVGKRVRYRVDGS-KIMKVFLDPKERN 161
RPPK+ S T + + +D G+ + ++V+ + +++KVF D ER
Sbjct: 122 RPPKRKSPDDNEATDNIQINFSIIDQD--------GRHMYFKVNHNLELIKVFKDFCERK 277
Query: 162 NTEYKLETFAAVYRKLSGK 180
N EY+ F + GK
Sbjct: 278 NLEYETMQFLCDGIHIKGK 334
>TC210001 similar to UP|Q9MA74 (Q9MA74) Histidine decarboxylase (Serine
decarboxylase), partial (42%)
Length = 791
Score = 26.9 bits (58), Expect = 5.8
Identities = 12/37 (32%), Positives = 23/37 (61%)
Frame = -2
Query: 101 ILRPPKKGSAAQRPRTRTLTAVHEAMLEDVVLPAEIV 137
I+RP K AA+ + R+ ++H+A+ E V +P ++
Sbjct: 466 IVRPLKHNGAAELIQHRSNASIHKAIFEVVCIPQTLL 356
>TC229365
Length = 1346
Score = 26.6 bits (57), Expect = 7.5
Identities = 11/22 (50%), Positives = 13/22 (59%)
Frame = +2
Query: 16 PTEFEETVGQALFDLENTHQEL 37
P +FE VGQ L + NTH L
Sbjct: 407 PLQFESPVGQLLEQISNTHPHL 472
>TC217580 similar to UP|Q84VX5 (Q84VX5) At4g02100, partial (74%)
Length = 2139
Score = 26.2 bits (56), Expect = 9.8
Identities = 16/59 (27%), Positives = 24/59 (40%)
Frame = +1
Query: 104 PPKKGSAAQRPRTRTLTAVHEAMLEDVVLPAEIVGKRVRYRVDGSKIMKVFLDPKERNN 162
PP +RPR R L H +LE+ PA+ + R + ++ F R N
Sbjct: 928 PPPPRRRTRRPRRRALLRGHPPLLENRRRPAQRAAELPRRMLHAPRLRAPFRRANRRVN 1104
>BF325009
Length = 382
Score = 26.2 bits (56), Expect = 9.8
Identities = 16/27 (59%), Positives = 19/27 (70%), Gaps = 3/27 (11%)
Frame = -2
Query: 97 ATRRILRPPKKGSAAQ-RPRT--RTLT 120
+TR L PPK GSAA+ RP + RTLT
Sbjct: 132 STRNFLSPPKTGSAARVRPPSSGRTLT 52
>TC206154 homologue to UP|FTSH_MEDSA (Q9BAE0) Cell division protein ftsH
homolog, chloroplast precursor , partial (88%)
Length = 2059
Score = 26.2 bits (56), Expect = 9.8
Identities = 16/46 (34%), Positives = 21/46 (44%)
Frame = +1
Query: 80 LVRELEKKFSGKDVILIATRRILRPPKKGSAAQRPRTRTLTAVHEA 125
L + K KD I A RI+ P+K +A + L A HEA
Sbjct: 1180 LAARRDLKEISKDEISDALERIIAGPEKKNAVVSDEKKKLVAYHEA 1317
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,618,725
Number of Sequences: 63676
Number of extensions: 55462
Number of successful extensions: 339
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 335
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 336
length of query: 191
length of database: 12,639,632
effective HSP length: 92
effective length of query: 99
effective length of database: 6,781,440
effective search space: 671362560
effective search space used: 671362560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0073.3


