

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0073.15
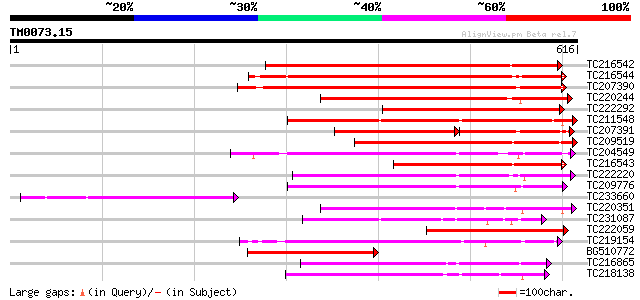
(616 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 362 e-100
TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 360 e-100
TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%) 334 6e-92
TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kina... 285 5e-77
TC222292 similar to UP|Q9FID5 (Q9FID5) Receptor protein kinase-l... 277 1e-74
TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kina... 271 8e-73
TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fra... 173 5e-67
TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Re... 249 2e-66
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 218 5e-57
TC216543 weakly similar to UP|Q84V32 (Q84V32) Receptor kinase LR... 218 6e-57
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 215 4e-56
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 214 7e-56
TC233660 211 6e-55
TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , pa... 192 4e-49
TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kin... 189 2e-48
TC222059 similar to GB|CAD39676.1|21741394|OSJN00121 OSJNBb0051N... 187 2e-47
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 181 1e-45
BG510772 weakly similar to GP|15408731|db P0443D08.13 {Oryza sat... 178 5e-45
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 177 1e-44
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 176 4e-44
>TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(38%)
Length = 1435
Score = 362 bits (929), Expect = e-100
Identities = 170/324 (52%), Positives = 242/324 (74%), Gaps = 2/324 (0%)
Frame = +2
Query: 279 WKAKKVD--QDIEAFIRNNGPQAIKRYSYSEIKKVTNSFKSKLGQGGYGQVYKGNLNNNC 336
W+ + + ++IE ++ N I YSY EIKK+ FK KLG GGYG V+KG L++
Sbjct: 302 WRKRHLSIYENIENYLEQNNLMPIG-YSYKEIKKMARGFKEKLGGGGYGIVFKGKLHSGP 478
Query: 337 PVAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKF 396
VA+K+L+ +KGNGQ+F++E+ +IGR H NVV L+G+C+EG K+AL+YEFMPNGSL+KF
Sbjct: 479 SVAIKMLHKAKGNGQDFISEIATIGRIHHQNVVQLIGYCVEGSKRALVYEFMPNGSLDKF 658
Query: 397 THKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADF 456
K+ +L+++ ++ IA G+A+G+ YLH GC +ILHFDIKP NILLD+ F PK++DF
Sbjct: 659 IFTKDGNIHLTYDEIYNIAIGVARGIAYLHHGCEMQILHFDIKPHNILLDETFTPKVSDF 838
Query: 457 GLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNI 516
GLAKL +SI++M ARGTIGY+APE++ +N GG+SHK+DVYS+GML++E+ ++N+
Sbjct: 839 GLAKLYPIDNSIVTMTAARGTIGYMAPELFYKNIGGISHKADVYSFGMLLMEMASKRKNL 1018
Query: 517 RNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPP 576
A SS+ YFP WIY + E+++ +G++ EEN+I KKM+IV LWCIQ P++RP
Sbjct: 1019NPHADHSSQLYFPFWIYNQLGKETDIEM-EGVTEEENKIAKKMIIVSLWCIQLKPTDRPS 1195
Query: 577 MSKVVEMLEGSIEQLQIPPKPFMF 600
M+KVVEMLEG IE L+IPPKP ++
Sbjct: 1196MNKVVEMLEGDIESLEIPPKPSLY 1267
>TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(36%)
Length = 1229
Score = 360 bits (925), Expect = e-100
Identities = 178/348 (51%), Positives = 247/348 (70%), Gaps = 2/348 (0%)
Frame = +2
Query: 260 ICITICIFRRKLSPIVSKHWKAKKVD--QDIEAFIRNNGPQAIKRYSYSEIKKVTNSFKS 317
I +T+ +F I++ W+ + + + IE ++ N I RYSY E+KK+ FK
Sbjct: 188 ILLTVSLF----IVILTCKWRKRHLSMFESIENYLEQNNLMPI-RYSYKEVKKMAGGFKD 352
Query: 318 KLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLE 377
KLG+GGYG V+KG L + VA+K+L SKGNGQ+F++EV +IGRT H N+V L+GFC+
Sbjct: 353 KLGEGGYGSVFKGKLGSGSCVAIKMLGKSKGNGQDFISEVATIGRTYHQNIVQLIGFCVH 532
Query: 378 GQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFD 437
G K+AL+YEFMPNGSL+KF K+ +LS++R++ I+ +A+G+ YLH GC +ILHFD
Sbjct: 533 GSKRALVYEFMPNGSLDKFIFSKDESIHLSYDRIYNISIEVARGIAYLHYGCEMQILHFD 712
Query: 438 IKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKS 497
IKP NILLD+NF PK++DFGLAKL +SI+ ARGTIGY+APE++ N GG+SHK+
Sbjct: 713 IKPHNILLDENFTPKVSDFGLAKLYPIDNSIVPRTAARGTIGYMAPELFYNNIGGISHKA 892
Query: 498 DVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEENEICK 557
DVYSYGML++E+ ++N+ A SS+ +FP WIY HI E ++ D + E E K
Sbjct: 893 DVYSYGMLLMEMASKRKNLNPHAERSSQLFFPFWIYNHIGDEEDIEMED---VTEEE--K 1057
Query: 558 KMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKPFMFSPTKT 605
KM+IV LWCIQ P++RP M+KVVEMLEG IE L+IPPKP ++ P++T
Sbjct: 1058KMIIVALWCIQLKPNDRPSMNKVVEMLEGDIENLEIPPKPTLY-PSET 1198
>TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%)
Length = 1342
Score = 334 bits (857), Expect = 6e-92
Identities = 170/362 (46%), Positives = 247/362 (67%), Gaps = 4/362 (1%)
Frame = +3
Query: 248 QGSIAGGIGALLI-CITICIFRRKLSPIVSKHWKAKKVDQD-IEAFIRNNGPQAIKRYSY 305
Q I G++L+ + I IF+ ++ +++ K+ DQ + F+ + + R++Y
Sbjct: 60 QSFIFATTGSILLGLVAIVIFK------IALYFRQKEEDQARVAKFLEDYRAEKPARFTY 221
Query: 306 SEIKKVTNSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSIGRTSH 365
+++K++T FK KLG+G +G V++G L+N VAVK+LN ++G G+EF+NEV +G+ H
Sbjct: 222 ADLKRITGGFKEKLGEGAHGAVFRGKLSNEILVAVKILNNTEGEGKEFINEVGIMGKIHH 401
Query: 366 VNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETN-LSWERLHKIAEGIAKGLEY 424
+NVV LLGFC EG +AL+Y PNGSL++F + + + L WE+L +IA GIAKG+EY
Sbjct: 402 INVVRLLGFCAEGFHRALVYNLFPNGSLQRFIVPPDDKDHFLGWEKLQQIALGIAKGIEY 581
Query: 425 LHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPE 484
LH+GCN I+HFDI P N+LLD NF PKI+DFGLAKLCS+ S++SM ARGT+GYIAPE
Sbjct: 582 LHEGCNQPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLVSMTAARGTLGYIAPE 761
Query: 485 VWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAW 544
V+++NFG VS+KSD+YSYGML+LE+VG ++N+ ++ +P WI+ I+ + ++
Sbjct: 762 VFSKNFGNVSYKSDIYSYGMLLLEMVGGRKNVDMSSAQDFHVLYPDWIHNLIDGDVHIHV 941
Query: 545 HDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE-GSIEQLQIPPKPFMFSPT 603
D I +I KK+ IVGLWCIQ P NRP + V++MLE G QL +PP PF S T
Sbjct: 942 EDECDI---KIAKKLAIVGLWCIQWQPVNRPSIKSVIQMLETGGESQLNVPPNPFQ-STT 1109
Query: 604 KT 605
T
Sbjct: 1110ST 1115
>TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kinase, partial
(32%)
Length = 1112
Score = 285 bits (728), Expect = 5e-77
Identities = 143/279 (51%), Positives = 197/279 (70%), Gaps = 5/279 (1%)
Frame = +1
Query: 338 VAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKF- 396
VAVK+LN + G+G++F+NEV ++G+ HVNVV LLGFC +G +AL+Y+F PNGSL++F
Sbjct: 40 VAVKILNDTVGDGKDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQRFL 219
Query: 397 THKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADF 456
N + L WE+L +IA G+AKG+EYLH GC+ RILHFDI P NIL+D +F PKI DF
Sbjct: 220 APPDNKDVFLGWEKLQQIALGVAKGVEYLHLGCDQRILHFDINPHNILIDDHFVPKITDF 399
Query: 457 GLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNI 516
GLAKLC + S +S+ ARGT+GYIAPEV++RNFG VS+KSD+YSYGML+LE+VG ++N
Sbjct: 400 GLAKLCPKNQSTVSITAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRKNT 579
Query: 517 RNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEEN---EICKKMVIVGLWCIQTIPSN 573
A +S + +P WI+ ++ ++IE+ I KK+ IVGLWCI+ P +
Sbjct: 580 NMSAEESFQVLYPEWIHNLLKSRD-----VQVTIEDEGDVRIAKKLAIVGLWCIEWNPID 744
Query: 574 RPPMSKVVEMLEGSIEQLQIPPKPF-MFSPTKTEVESGT 611
RP M V++MLEG ++L PP PF S ++T V + T
Sbjct: 745 RPSMKTVIQMLEGDGDKLIAPPTPFDKTSSSRTSVVAPT 861
>TC222292 similar to UP|Q9FID5 (Q9FID5) Receptor protein kinase-like protein,
partial (16%)
Length = 732
Score = 277 bits (708), Expect = 1e-74
Identities = 130/197 (65%), Positives = 154/197 (77%)
Frame = +2
Query: 406 LSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSET 465
LSW+ +IA GIA+GLEYLH+GCNTRILHFDIKP NILLD+NFCPKI+DFGLAKLC
Sbjct: 2 LSWDNFWQIAIGIARGLEYLHRGCNTRILHFDIKPHNILLDENFCPKISDFGLAKLCPRK 181
Query: 466 HSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSE 525
SIISM D RGTIGY+APEVWNRNFGGVSHKSDVY YGM++LE+VG + NI EAS +SE
Sbjct: 182 GSIISMSDPRGTIGYVAPEVWNRNFGGVSHKSDVYRYGMMLLEMVGGRNNINAEASHTSE 361
Query: 526 TYFPHWIYKHIEVESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLE 585
YFP WIYK +E +L + M+ EENEI K+M +VGLWC+QT P +RP M++VV+MLE
Sbjct: 362 IYFPDWIYKRLEQGGDLRPNGVMAPEENEIVKRMPVVGLWCVQTFPKDRPAMTRVVDMLE 541
Query: 586 GSIEQLQIPPKPFMFSP 602
G + L IP F P
Sbjct: 542 GKMNSLDIPKNLFFLLP 592
>TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kinase
TAK33-like protein, partial (58%)
Length = 1194
Score = 271 bits (692), Expect = 8e-73
Identities = 146/323 (45%), Positives = 212/323 (65%), Gaps = 8/323 (2%)
Frame = +2
Query: 302 RYSYSEIKKVTNSFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNAS--KGNGQEFLNEVVS 359
R++ +++ T+++ LG GG+G VYKG+ +N VAVKVL S K ++F+ EV +
Sbjct: 11 RFTDQQLRIATDNYSFLLGSGGFGVVYKGSFSNGTIVAVKVLRGSSDKRIDEQFMAEVGT 190
Query: 360 IGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIA 419
IG+ H N+V L GFC E +AL+YE+M NG+LEK+ +N T LS+E+LH+IA G A
Sbjct: 191 IGKVHHFNLVRLYGFCFERHLRALVYEYMVNGALEKYLFHEN--TTLSFEKLHEIAVGTA 364
Query: 420 KGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIG 479
+G+ YLH+ C RI+H+DIKP NILLD+NFCPK+ADFGLAKLC+ ++ ISM RGT G
Sbjct: 365 RGIAYLHEECQQRIIHYDIKPGNILLDRNFCPKVADFGLAKLCNRDNTHISMTGGRGTPG 544
Query: 480 YIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVE 539
Y APE+W V+HK DVYS+GML+ EI+G ++N N S+ +FP W+++ + E
Sbjct: 545 YAAPELWLP--FPVTHKCDVYSFGMLLFEIIGRRRN-HNINLPESQVWFPMWVWERFDAE 715
Query: 540 S--NLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKP 597
+ +L G+ + EI +++V V L C+Q P RP MS VV+MLEGS+E + P P
Sbjct: 716 NVEDLISACGIEDQNREIAERIVKVALSCVQYKPEARPIMSVVVKMLEGSVE-VPKPLNP 892
Query: 598 FM----FSPTKTEVESGTTSNSD 616
F ++P T+ + +N+D
Sbjct: 893 FQHLIDWTPPPTDPVQASQTNTD 961
>TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fragment),
partial (29%)
Length = 963
Score = 173 bits (438), Expect(2) = 5e-67
Identities = 82/137 (59%), Positives = 104/137 (75%), Gaps = 1/137 (0%)
Frame = +1
Query: 353 FLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETN-LSWERL 411
F+NEV +G+ H+NVV LLG+C EG +AL+Y F PNGSL+ F + + N L WE+L
Sbjct: 19 FINEVEIMGKIHHINVVRLLGYCAEGIHRALVYNFFPNGSLQSFIFPPDDKQNFLGWEKL 198
Query: 412 HKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISM 471
IA GIAKG+ YLH+GCN I+HFDI P N+LLD NF PKI+DFGLAKLCS+ S++SM
Sbjct: 199 QNIALGIAKGIGYLHQGCNHPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLVSM 378
Query: 472 HDARGTIGYIAPEVWNR 488
ARGT+GYIAPEV++R
Sbjct: 379 TAARGTLGYIAPEVFSR 429
Score = 100 bits (249), Expect(2) = 5e-67
Identities = 53/126 (42%), Positives = 78/126 (61%), Gaps = 1/126 (0%)
Frame = +2
Query: 489 NFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAWHDGM 548
NFG VS+KSD+YSYGML+LE+VG ++N+ + + +P W++ + + ++ D
Sbjct: 431 NFGNVSYKSDIYSYGMLLLEMVGGRKNVDTSSPEDFHVLYPDWMHDLVHGDVHIHVEDEG 610
Query: 549 SIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQ-LQIPPKPFMFSPTKTEV 607
+ +I +K+ IVGLWCIQ P NRP + V++MLE E L +PP PF S T T +
Sbjct: 611 DV---KIARKLAIVGLWCIQWQPLNRPSIKSVIQMLESKEEDLLTVPPNPF-HSSTST-I 775
Query: 608 ESGTTS 613
SG TS
Sbjct: 776 PSGFTS 793
>TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Receptor
kinase LRK14), partial (19%)
Length = 866
Score = 249 bits (637), Expect = 2e-66
Identities = 121/242 (50%), Positives = 168/242 (69%)
Frame = +2
Query: 375 CLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRIL 434
C EG+K AL+YEFMPNGSL+K+ K +LS+++ ++I GIA+G+ YLH+ C+ +IL
Sbjct: 2 CAEGEKHALVYEFMPNGSLDKYIFSKEESVSLSYDKTYEICLGIARGIAYLHQDCDVQIL 181
Query: 435 HFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVS 494
HFDIKP NILLD NF PK++DFGLAKL I + RGT GY+APE++ +N GGVS
Sbjct: 182 HFDIKPHNILLDDNFIPKVSDFGLAKLYPIKDKSIILTGLRGTFGYMAPELFYKNIGGVS 361
Query: 495 HKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEENE 554
+K+DVYS+GML++E+ ++N SS+ +FP WIY H E ++ + +S E+
Sbjct: 362 YKADVYSFGMLLMEMGSRRRNSNPHTEHSSQHFFPFWIYDHFMEEKDIHMEE-VSEEDKI 538
Query: 555 ICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKPFMFSPTKTEVESGTTSN 614
+ KKM IV LWCIQ P++RP M KVVEMLEG +E + +PPKP +F P +T ++S S
Sbjct: 539 LVKKMFIVSLWCIQLKPNDRPSMKKVVEMLEGKVENIDMPPKP-VFYPHETTIDSDQASW 715
Query: 615 SD 616
SD
Sbjct: 716 SD 721
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 218 bits (556), Expect = 5e-57
Identities = 144/398 (36%), Positives = 209/398 (52%), Gaps = 24/398 (6%)
Frame = +3
Query: 241 STTSNNCQGSIAGGIGALLICIT---------ICIFRRKLSPIVSKHWKAKKVDQDIEAF 291
S S G AG I A+++ IT IC R+ K K DI
Sbjct: 600 SPISPGSSGISAGTIVAIVVPITVAVLIFIVGICFLSRRARKKQQGSVKEGKTAYDIPTV 779
Query: 292 IRNNGPQAIKRYSYSEIKKVTNSFKS--KLGQGGYGQVYKGNLNNNCPVAVKVLNASKGN 349
++ +S I+ TN F + KLG+GG+G+VYKG L++ VAVK L+ S G
Sbjct: 780 DS-------LQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQ 938
Query: 350 G-QEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSW 408
G +EF NEVV + + H N+V LLGFCL+G++K L+YE++PN SL+ + L W
Sbjct: 939 GGEEFKNEVVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDW 1118
Query: 409 ERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSI 468
R +KI GIA+G++YLH+ RI+H D+K SNILLD + PKI+DFG+A++ +
Sbjct: 1119GRRYKIIGGIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQ 1298
Query: 469 ISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYF 528
+ GT GY+APE G S KSDVYS+G+L++EI+ K+N +D +E
Sbjct: 1299GNTSRIVGTYGYMAPEY--AMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLL 1472
Query: 529 PH-WIYKHIEVESNLAWHDGMSIE-----------ENEICKKMVIVGLWCIQTIPSNRPP 576
+ W W DG +E +NE+ + + +GL C+Q P++RP
Sbjct: 1473SYAW----------QLWKDGTPLELMDPILRESYNQNEVIRS-IHIGLLCVQEDPADRPT 1619
Query: 577 MSKVVEMLEGSIEQLQIPPKPFMFSPTKTEVESGTTSN 614
M+ +V ML+ + L P +P F V SGT N
Sbjct: 1620MATIVLMLDSNTVTLPTPTQPAFF------VHSGTDPN 1715
>TC216543 weakly similar to UP|Q84V32 (Q84V32) Receptor kinase LRK45, partial
(23%)
Length = 603
Score = 218 bits (555), Expect = 6e-57
Identities = 103/188 (54%), Positives = 140/188 (73%)
Frame = +2
Query: 418 IAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGT 477
+A+G+ YLH GC +ILHFDIKP N LD+NF PK++DFGLAKL +SI+ ARGT
Sbjct: 2 VARGIAYLHYGCEMQILHFDIKPHNXXLDENFTPKVSDFGLAKLYPIDNSIVPRTAARGT 181
Query: 478 IGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIE 537
IGY+APE++ N GG+SHK+DVYSYGML++E+ ++N+ A SS+ +FP WIY HI
Sbjct: 182 IGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKRKNLNPHAERSSQLFFPFWIYNHIG 361
Query: 538 VESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKP 597
E ++ D ++ EE ++ KKM+IV LWCIQ P++RP M+KVVEMLEG IE L+IPPKP
Sbjct: 362 DEEDIEMED-VTEEEKKMVKKMIIVALWCIQLKPNDRPSMNKVVEMLEGDIENLEIPPKP 538
Query: 598 FMFSPTKT 605
++ P++T
Sbjct: 539 TLY-PSET 559
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 215 bits (548), Expect = 4e-56
Identities = 124/315 (39%), Positives = 184/315 (58%), Gaps = 8/315 (2%)
Frame = +3
Query: 308 IKKVTN--SFKSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRTS 364
I+ TN S++ ++G+GG+G VYKG L + +AVK L+ S G G EF NE++ I +
Sbjct: 204 IEAATNKFSYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIAKLQ 383
Query: 365 HVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEY 424
H N+V LLGFCLE +K LIYEF+ N SL+ F + L+W +KI EGIA+G+ Y
Sbjct: 384 HRNLVTLLGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQLNWSERYKIIEGIAQGISY 563
Query: 425 LHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPE 484
LH+ +++H D+KPSN+LLD N PKI+DFG+A++ + + GT GY++PE
Sbjct: 564 LHEHSRLKVIHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIVGTYGYMSPE 743
Query: 485 VWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEVESNLAW 544
G S KSDV+S+G+++LEI+ AK+N R+ SD + W E N+
Sbjct: 744 Y--AMHGQFSEKSDVFSFGVIVLEIISAKRNTRSVFSDHDDLLSYTWEQWMDEAPLNIF- 914
Query: 545 HDGMSIEENEICK-----KMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKPFM 599
SIE E C K + +GL C+Q P +RP +++V+ L SI +L +P KP
Sbjct: 915 --DQSIEA-EFCDHSEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLPKKPIR 1085
Query: 600 FSPTKTEVESGTTSN 614
S ++ G +S+
Sbjct: 1086QSGIVQKIAVGESSS 1130
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 214 bits (546), Expect = 7e-56
Identities = 125/311 (40%), Positives = 187/311 (59%), Gaps = 6/311 (1%)
Frame = +2
Query: 302 RYSYSEIKKVTNSFK--SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVV 358
R+ +S I+ T F +KLG+GG+G+VYKG L + VAVK L+ G G +EF NEV
Sbjct: 41 RFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEEFKNEVE 220
Query: 359 SIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGI 418
+ + H N+V LLGFCLEG++K L+YEF+ N SL+ + +L W R +KI EGI
Sbjct: 221 IVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVEGI 400
Query: 419 AKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTI 478
A+G++YLH+ +I+H D+K SN+LLD + PKI+DFG+A++ + + + GT
Sbjct: 401 ARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVGTY 580
Query: 479 GYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQNIRNEASDSSETYFPHWIYKHIEV 538
GY++PE G S KSDVYS+G+LILEI+ K+N +D +E + +K +
Sbjct: 581 GYMSPEY--AMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLL-SYAWKLWKD 751
Query: 539 ESNLAWHDGM---SIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPP 595
E+ L D S NE+ + + +GL C+Q P +RP M+ VV ML+ LQ+P
Sbjct: 752 EAPLELMDQSLRESYTRNEVI-RCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVTLQVPN 928
Query: 596 KPFMFSPTKTE 606
+P + ++TE
Sbjct: 929 QPAFYINSRTE 961
>TC233660
Length = 1039
Score = 211 bits (538), Expect = 6e-55
Identities = 108/241 (44%), Positives = 142/241 (58%), Gaps = 4/241 (1%)
Frame = +3
Query: 12 ALFLCFIISSGSH--NTSSSTTCPAYRCTNGPNISYPFWLARGSPPDQYCGYQELGLICY 69
ALF II H ++STTC +C N NISYPFW + ++CGY E GL C
Sbjct: 15 ALFALSIILVTCHAQTLNTSTTCEPSKCGN-LNISYPFWKKSNTNVQEFCGYPEFGLECL 191
Query: 70 DGDPIFSLPPGLYYYVKDIDYENHSLKLVDADTANQTCPRALHNVPVGNLPLSHSPLNKN 129
D I P Y V DI+Y+ HS+ L+D D Q CPRA HNV + NLPLS S L+ N
Sbjct: 192 DDQAIMIFPTDRYQ-VTDINYDIHSITLLDIDVLGQPCPRARHNVSLHNLPLSFSSLDFN 368
Query: 130 LSFYYNCSGYPSGVPFIECLSSGVNRSFVFEMGN--ETKGFDWDENCQVNVVVTVMKDEV 187
LSFY+NCS YPS + I C+ +S+VF+ G+ E+ GFDW +C+ +VVVTV +DE+
Sbjct: 369 LSFYFNCSSYPSSIQHIGCMEHDKYQSYVFKTGDEAESNGFDWLRHCEEHVVVTVKQDEI 548
Query: 188 TSDGLMSEFAGAMNEGFVLDWQTPASCAECEASDGVCGYSNTKKELLCFCKDGSTTSNNC 247
L++ F AM +GFVLDW CA CE S+G C + K+ C C DG T + +C
Sbjct: 549 EISSLITGFGDAMQKGFVLDWMRAQDCAVCEESNGYCRFDQATKQSRCLCSDGRTEAKSC 728
Query: 248 Q 248
+
Sbjct: 729 K 731
>TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , partial (11%)
Length = 1064
Score = 192 bits (488), Expect = 4e-49
Identities = 116/289 (40%), Positives = 170/289 (58%), Gaps = 11/289 (3%)
Frame = +1
Query: 338 VAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFT 397
VAVK L + ++F EV +I T H+N+V L+GFC EG+ + L+YEFM NGSL+ F
Sbjct: 1 VAVKQLEGIEQGEKQFRMEVATISSTHHLNLVRLIGFCSEGRHRLLVYEFMKNGSLDDFL 180
Query: 398 HKKNFETN--LSWERLHKIAEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIAD 455
+ L+WE IA G A+G+ YLH+ C I+H DIKP NILLD+N+ K++D
Sbjct: 181 FLTEQHSGKLLNWEYRFNIALGTARGITYLHEECRDCIVHCDIKPENILLDENYVAKVSD 360
Query: 456 FGLAKLCS-ETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAKQ 514
FGLAKL + + H ++ RGT GY+APE W N ++ KSDVY YGM++LEIV ++
Sbjct: 361 FGLAKLINPKDHRHRTLTSVRGTRGYLAPE-WLANL-PITSKSDVYGYGMVLLEIVSGRR 534
Query: 515 NIRNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEENEI----CKKMVIVGLWCIQTI 570
N + + +++ F W Y+ E + N++ + E+ ++ + WCIQ
Sbjct: 535 NF-DVSEETNRKKFSIWAYEEFE-KGNISGILDKRLANQEVDMEQVRRAIQASFWCIQEQ 708
Query: 571 PSNRPPMSKVVEMLEGSIEQLQIP-PKPFM---FSPTKTEVESGTTSNS 615
PS+RP MS+V++MLEG E + P PK M S T T + S ++ S
Sbjct: 709 PSHRPTMSRVLQMLEGVTEPERPPAPKSVMEGAVSGTSTYLSSNASAFS 855
>TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kinase
precursor, partial (30%)
Length = 826
Score = 189 bits (481), Expect = 2e-48
Identities = 112/284 (39%), Positives = 164/284 (57%), Gaps = 19/284 (6%)
Frame = +1
Query: 319 LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLEG 378
+G GG+G V++G L++ VAVK L G +EF EV +IG HVN+V L GFC E
Sbjct: 1 VGHGGFGTVFQGELSDASVVAVKRLERPGGGEKEFRAEVSTIGNIQHVNLVRLRGFCSEN 180
Query: 379 QKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRILHFDI 438
+ L+YE+M NG+L + K+ LSW+ ++A G AKG+ YLH+ C I+H DI
Sbjct: 181 SHRLLVYEYMQNGALNVYLRKEG--PCLSWDVRFRVAVGTAKGIAYLHEECRCCIIHCDI 354
Query: 439 KPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSD 498
KP NILLD +F K++DFGLAKL S + + RGT GY+APE W ++ K+D
Sbjct: 355 KPENILLDGDFTAKVSDFGLAKLIGRDFSRV-LVTMRGTWGYVAPE-WISGV-AITTKAD 525
Query: 499 VYSYGMLILEIVGAKQNIR-----------NEASD--SSETYFPHWIYKHIEVESNLA-- 543
VYSYGM +LE++G ++N+ E+ D + +FP W + I +E N++
Sbjct: 526 VYSYGMTLLELIGGRRNVEAPLSAGGGGGGGESGDEMGGKWFFPPWAAQRI-IEGNVSDV 702
Query: 544 ----WHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEM 583
+ +IEE +++ +V +WCIQ + RP M VV+M
Sbjct: 703 MDKRLGNAYNIEE---ARRVALVAVWCIQDDEAMRPTMGMVVKM 825
>TC222059 similar to GB|CAD39676.1|21741394|OSJN00121 OSJNBb0051N19.7 {Oryza
sativa (japonica cultivar-group);} , partial (30%)
Length = 510
Score = 187 bits (474), Expect = 2e-47
Identities = 89/154 (57%), Positives = 115/154 (73%)
Frame = +2
Query: 454 ADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIVGAK 513
+DFGLAK+C+ S+IS+ ARGT GYIAPEV++RNFG VSHKSDVYSYGM+ILE+VG +
Sbjct: 2 SDFGLAKICTRKESMISIFGARGTAGYIAPEVFSRNFGTVSHKSDVYSYGMMILEMVGRR 181
Query: 514 QNIRNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEENEICKKMVIVGLWCIQTIPSN 573
+NI+ E + SSE YFP WIY +E L + + ++++ +KM IVGLWCIQT PS
Sbjct: 182 KNIKTEVNCSSEIYFPDWIYNRLESNEELGLQNIRNESDDKLVRKMTIVGLWCIQTHPST 361
Query: 574 RPPMSKVVEMLEGSIEQLQIPPKPFMFSPTKTEV 607
RP +SKV+EML +E LQIPPKPF+ SP + V
Sbjct: 362 RPAISKVLEMLGSKVELLQIPPKPFLSSPPTSPV 463
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 181 bits (458), Expect = 1e-45
Identities = 126/360 (35%), Positives = 192/360 (53%), Gaps = 9/360 (2%)
Frame = +1
Query: 250 SIAGGIGALLICITIC-IFRRKLSPIVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEI 308
S+A +G +L IC I+RR I K K +D+ ++ + + I
Sbjct: 190 SVAAPLGVVL---AICFIYRRN---IADKSKTKKSIDRQLQDV-------DVPLFDMLTI 330
Query: 309 KKVTNSF--KSKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRTSH 365
T++F +K+G+GG+G VYKG L +AVK L++ G G EF+ EV I + H
Sbjct: 331 TAATDNFLLNNKIGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQH 510
Query: 366 VNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKNFETNLSWERLHKIAEGIAKGLEYL 425
N+V LLG C++GQ+K L+YE++ NGSL F + L W R I GIA+GL YL
Sbjct: 511 RNLVKLLGCCIKGQEKLLVYEYVVNGSLNSFIFDQIKSKLLDWPRRFNIILGIARGLLYL 690
Query: 426 HKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPE- 484
H+ RI+H D+K SN+LLD+ PKI+DFG+A+ + + + GT GY+APE
Sbjct: 691 HQDSRLRIIHRDLKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTNRVVGTYGYMAPEY 870
Query: 485 VWNRNFGGVSHKSDVYSYGMLILEIVGAKQN----IRNEASDSSETYFPHWIYKHIEVES 540
++ NF S KSDV+S+G+L+LEIV +N N+ + + W ++
Sbjct: 871 AFDGNF---SIKSDVFSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWALWKEQNALQLI 1041
Query: 541 NLAWHDGMSIEENEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSIEQLQIPPKPFMF 600
+ D I E C + V L C+Q P +RP M+ V++ML ++ ++ P +P F
Sbjct: 1042DSGIKDSCVIPEVLRC---IHVSLLCVQQYPEDRPTMTSVIQMLGSEMDMVE-PKEPGFF 1209
>BG510772 weakly similar to GP|15408731|db P0443D08.13 {Oryza sativa
(japonica cultivar-group)}, partial (16%)
Length = 437
Score = 178 bits (452), Expect = 5e-45
Identities = 88/143 (61%), Positives = 111/143 (77%), Gaps = 1/143 (0%)
Frame = +1
Query: 259 LICITICIFRRKLSP-IVSKHWKAKKVDQDIEAFIRNNGPQAIKRYSYSEIKKVTNSFKS 317
LI + IC + ++ I+ + K KK DQ IEAF+ + G +KRYS+S+IKK+TNSFK
Sbjct: 1 LIAVIICRNKARIWKFILVQVGKIKKNDQVIEAFLESQGSLGLKRYSFSDIKKITNSFKI 180
Query: 318 KLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQEFLNEVVSIGRTSHVNVVNLLGFCLE 377
KLG+GGYG VYKG L N C VAVK+LN SK NG+EF+NEV SI +TSHVN+V+LLGFCL+
Sbjct: 181 KLGEGGYGSVYKGKLLNGCSVAVKILNESKENGEEFINEVASISKTSHVNIVSLLGFCLD 360
Query: 378 GQKKALIYEFMPNGSLEKFTHKK 400
G +KALIYEFM NGSLEK+ H+K
Sbjct: 361 GSRKALIYEFMSNGSLEKYIHEK 429
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 177 bits (449), Expect = 1e-44
Identities = 107/276 (38%), Positives = 159/276 (56%), Gaps = 4/276 (1%)
Frame = +1
Query: 317 SKLGQGGYGQVYKGNLNNNCPVAVKVLNASKGNG-QEFLNEVVSIGRTSHVNVVNLLGFC 375
+K+G+GG+G VYKG ++ +AVK L++ G +EFLNE+ I H ++V L G C
Sbjct: 7 NKIGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCC 186
Query: 376 LEGQKKALIYEFMPNGSLEKFTH-KKNFETNLSWERLHKIAEGIAKGLEYLHKGCNTRIL 434
+EG + L+YE+M N SL + + + L W +KI GIA+GL YLH+ +I+
Sbjct: 187 VEGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIV 366
Query: 435 HFDIKPSNILLDKNFCPKIADFGLAKLCSETHSIISMHDARGTIGYIAPEVWNRNFGGVS 494
H DIK +N+LLD++ PKI+DFGLAKL E ++ IS A GT GY+APE G ++
Sbjct: 367 HRDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHISTRIA-GTFGYMAPEY--AMHGYLT 537
Query: 495 HKSDVYSYGMLILEIVGAKQNI--RNEASDSSETYFPHWIYKHIEVESNLAWHDGMSIEE 552
K+DVYS+G++ LEI+ + N R + S + H + + ++ + G+ +
Sbjct: 538 DKADVYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNK 717
Query: 553 NEICKKMVIVGLWCIQTIPSNRPPMSKVVEMLEGSI 588
E M+ V L C + RP MS VV MLEG I
Sbjct: 718 EE-ALVMIKVALLCTNVTAALRPTMSSVVSMLEGKI 822
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 176 bits (445), Expect = 4e-44
Identities = 111/298 (37%), Positives = 170/298 (56%), Gaps = 11/298 (3%)
Frame = +1
Query: 300 IKRYSYSEIKKVTNSFKSK--LGQGGYGQVYKGNLNNNCPVAVKVLNASKGNGQE--FLN 355
+KR+S E++ T+SF +K LG+GG+G+VYKG L + VAVK L + G E F
Sbjct: 112 LKRFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQT 291
Query: 356 EVVSIGRTSHVNVVNLLGFCLEGQKKALIYEFMPNGSLEKFTHKKN-FETNLSWERLHKI 414
EV I H N++ L GFC+ ++ L+Y +M NGS+ ++ ++ L W ++
Sbjct: 292 EVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPPYQEPLDWPTRKRV 471
Query: 415 AEGIAKGLEYLHKGCNTRILHFDIKPSNILLDKNFCPKIADFGLAKLC--SETHSIISMH 472
A G A+GL YLH C+ +I+H D+K +NILLD+ F + DFGLAKL +TH ++
Sbjct: 472 ALGSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAV- 648
Query: 473 DARGTIGYIAPEVWNRNFGGVSHKSDVYSYGMLILEIV-GAKQNIRNEASDSSETYFPHW 531
RGTIG+IAPE + G S K+DV+ YG+++LE++ G + ++ + W
Sbjct: 649 --RGTIGHIAPEYLST--GKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDW 816
Query: 532 IYKHIEVESNLAWHDGMSIEENEI---CKKMVIVGLWCIQTIPSNRPPMSKVVEMLEG 586
+ K + E L ++ N I ++++ V L C Q P +RP MS+VV MLEG
Sbjct: 817 V-KGLLKEKKLEMLVDPDLQTNYIETEVEQLIQVALLCTQGSPMDRPKMSEVVRMLEG 987
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,545,207
Number of Sequences: 63676
Number of extensions: 574071
Number of successful extensions: 4800
Number of sequences better than 10.0: 952
Number of HSP's better than 10.0 without gapping: 3917
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3965
length of query: 616
length of database: 12,639,632
effective HSP length: 103
effective length of query: 513
effective length of database: 6,081,004
effective search space: 3119555052
effective search space used: 3119555052
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0073.15


