

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
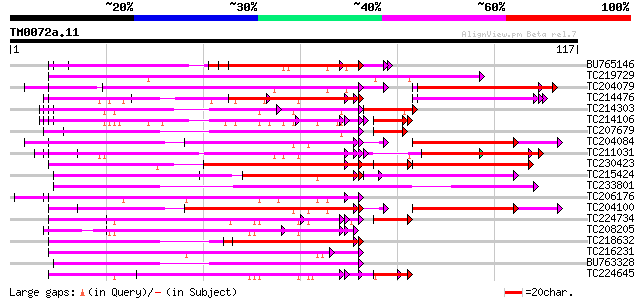
Query= TM0072a.11
(117 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 69 4e-13
TC219729 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 62 3e-11
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 53 8e-11
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 61 9e-11
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 54 2e-10
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 54 4e-10
TC207679 similar to UP|Q7XJI7 (Q7XJI7) Glycine-rich protein TomR... 59 4e-10
TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, co... 50 8e-10
TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 57 2e-09
TC230423 similar to UP|Q41187 (Q41187) Glycine-rich protein (Fra... 55 2e-09
TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%) 55 4e-09
TC233801 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 55 6e-09
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 54 1e-08
TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, ... 49 1e-08
TC224734 homologue to UP|Q09085 (Q09085) Hydroxyproline-rich gly... 53 2e-08
TC208205 hydroxyproline-rich glycoprotein 53 2e-08
TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wal... 52 3e-08
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 52 3e-08
BU763328 similar to SP|P10496|GRP2_ Glycine-rich cell wall struc... 52 3e-08
TC224645 homologue to UP|Q41707 (Q41707) Extensin class 1 protei... 52 3e-08
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri
f. nagariensis}, partial (25%)
Length = 407
Score = 68.6 bits (166), Expect = 4e-13
Identities = 35/65 (53%), Positives = 35/65 (53%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG EG GG GGGGGGG GGG G G GGG
Sbjct: 21 GGGGGGGGGGGGGGGGGGGGGGG---------EGGGGGGGGGGGGGGGGGGGGGRGGGGG 173
Query: 69 GGCGG 73
GG GG
Sbjct: 174GGGGG 188
Score = 67.0 bits (162), Expect = 1e-12
Identities = 34/65 (52%), Positives = 35/65 (53%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG+ GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 42 GGGGGGGGGGGGGGGGEGGGGGG----------GGGGGGGGGGGGGGGRGGGGGGGGGGG 191
Query: 69 GGCGG 73
GG GG
Sbjct: 192GGGGG 206
Score = 66.6 bits (161), Expect = 2e-12
Identities = 35/65 (53%), Positives = 37/65 (56%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG +GG G G GG GGGG V E GG+ GGGGGGG GGG G G GGG
Sbjct: 142 GGGGEGGGGGGGGGGGGGGGGGGGVF*GG---ERXREGGERGGGGGGGGGGGGGGGXGGG 312
Query: 69 GGCGG 73
GG GG
Sbjct: 313 GGGGG 327
Score = 66.2 bits (160), Expect = 2e-12
Identities = 35/65 (53%), Positives = 35/65 (53%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGGV EG GG GGGGGGG GGG G G GGG
Sbjct: 145 GGGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGGERGGGGGGGGGGGGGGGXGGGGGGG 324
Query: 69 GGCGG 73
G GG
Sbjct: 325 GRGGG 339
Score = 66.2 bits (160), Expect = 2e-12
Identities = 34/65 (52%), Positives = 35/65 (53%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G+ GG GGGGGGG GGG G G GGG
Sbjct: 22 GGGGGGGGGGGGGGGGGGGGGGGR--------GGEGGGGGGGGGGGGGGGGGEGGGGGGG 177
Query: 69 GGCGG 73
GG GG
Sbjct: 178GGGGG 192
Score = 65.1 bits (157), Expect = 5e-12
Identities = 34/65 (52%), Positives = 34/65 (52%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGGGGG GGG G G GGG
Sbjct: 32 GGGGGGGGGGGGGGGGGGGGGGG-------RGGGGGGGGGGGGGGGGGKGGGGGGGGGGG 190
Query: 69 GGCGG 73
GG GG
Sbjct: 191GGGGG 205
Score = 63.9 bits (154), Expect = 1e-11
Identities = 34/70 (48%), Positives = 35/70 (49%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGGG G GG GGGG GG GGG G G GGG
Sbjct: 51 GGGGGGGGGGGGGEGGGGGGGGG----------GGGGGGGGGGGGRGGGGGGGGGGGGGG 200
Query: 69 GGCGGLMWWR 78
GG GG + R
Sbjct: 201 GGGGGFLGGR 230
Score = 63.2 bits (152), Expect = 2e-11
Identities = 33/65 (50%), Positives = 34/65 (51%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G G GG GGG E+ G GG GGGGGGG GGG G G GG
Sbjct: 149 GGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEGG 328
Query: 69 GGCGG 73
GG GG
Sbjct: 329 GGGGG 343
Score = 62.8 bits (151), Expect = 2e-11
Identities = 34/69 (49%), Positives = 36/69 (51%), Gaps = 4/69 (5%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGG----DGGGNGCG 64
GG GG G G GG GGGG + + G GG GGGGGGG GGG G G
Sbjct: 147 GGGRGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGGGRG 326
Query: 65 SGGGGGCGG 73
GGGGG GG
Sbjct: 327 EGGGGGGGG 353
Score = 62.0 bits (149), Expect = 4e-11
Identities = 36/81 (44%), Positives = 38/81 (46%), Gaps = 10/81 (12%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGG----------GDG 58
GG GG G G GG GGGG + G GG GGGGGG G+G
Sbjct: 146 GGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEG 325
Query: 59 GGNGCGSGGGGGCGGLMWWRR 79
GG G G GGGGG GG RR
Sbjct: 326 GGGGGGGGGGGGGGGGRGGRR 388
Score = 56.6 bits (135), Expect = 2e-09
Identities = 30/65 (46%), Positives = 32/65 (49%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG +GG G G GG G GG G G+ GGGGGGG GGG G G GGG
Sbjct: 240 GGGGEGGGGGGGGGGGGGGGXGG----------GGGGRGEGGGGGGGGGGGGGGGGEGGG 389
Query: 69 GGCGG 73
GG
Sbjct: 390 RRGGG 404
Score = 54.3 bits (129), Expect = 8e-09
Identities = 30/61 (49%), Positives = 31/61 (50%)
Frame = +1
Query: 13 DGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCG 72
+GG G G GG GGGG G GG GGGGGGG GGG G G GGG G
Sbjct: 241 EGGERGGGGGGGGGGGGGGGX------GGGGGGGGRGGGGGGGGGGGGGGEGREEGGGGG 402
Query: 73 G 73
G
Sbjct: 403 G 405
Score = 52.0 bits (123), Expect = 4e-08
Identities = 30/64 (46%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Frame = +2
Query: 10 GFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCG----S 65
G GG G G GG GGGG G GG GGGGGGG GGG G G
Sbjct: 245 GGRGGGGGGGGGGGGGGGGGGG----------GGGEGGGGGGGGGGGGGGGGGRGGRREE 394
Query: 66 GGGG 69
GGGG
Sbjct: 395 GGGG 406
Score = 48.1 bits (113), Expect = 6e-07
Identities = 22/31 (70%), Positives = 22/31 (70%), Gaps = 1/31 (3%)
Frame = +1
Query: 44 CSGGDSGGGGGGGDGGGNGCGSGGG-GGCGG 73
C GG GGGGGGG GGG G G GGG GG GG
Sbjct: 16 CPGGGGGGGGGGGGGGGGGGGGGGGRGGEGG 108
Score = 45.8 bits (107), Expect = 3e-06
Identities = 21/32 (65%), Positives = 22/32 (68%)
Frame = +1
Query: 42 GDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G+ G GGGGGGG GGG G G GGGGG GG
Sbjct: 7 GEVCPGGGGGGGGGGGGGGGG-GGGGGGGRGG 99
Score = 43.9 bits (102), Expect = 1e-05
Identities = 19/28 (67%), Positives = 20/28 (70%)
Frame = +1
Query: 46 GGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG+ GGGGG GGG G G GGGGG GG
Sbjct: 4 GGEVCPGGGGGGGGGGGGGGGGGGGGGG 87
Score = 42.7 bits (99), Expect = 2e-05
Identities = 19/28 (67%), Positives = 19/28 (67%)
Frame = +3
Query: 46 GGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG GGGGG GGG G G GGGGG GG
Sbjct: 3 GGRGLPGGGGGGGGGGGGGGGGGGGGGG 86
Score = 31.6 bits (70), Expect = 0.056
Identities = 18/49 (36%), Positives = 20/49 (40%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD 57
GG G G G GG GGGG GG+ GG GGG+
Sbjct: 303 GGGGGGRGEGGGGGGGGGGGGGG--------------GGEGGGRRGGGE 407
>TC219729 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (7%)
Length = 785
Score = 62.4 bits (150), Expect = 3e-11
Identities = 44/97 (45%), Positives = 49/97 (50%), Gaps = 7/97 (7%)
Frame = -2
Query: 9 GGFADGGSSGDDGCGGDSD---GGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGS 65
GGF GG S G GG S GGGG +I G+ GGD G GGGG GGG+ G
Sbjct: 361 GGFGGGGHSTTTGGGGGSGSGFGGGGHLITGGGGDGGNGGGGDGGIGGGGDGGGGHSIGL 182
Query: 66 GGGGGCGGL---MWWRRRW-WRVVVVTSTVVATAVVT 98
GGGGG G L R W + V V TV + VVT
Sbjct: 181 GGGGGGGSLHSSHIMRGTWCFSSVEVDETVEGSTVVT 71
Score = 27.7 bits (60), Expect = 0.81
Identities = 21/58 (36%), Positives = 23/58 (39%), Gaps = 5/58 (8%)
Frame = -2
Query: 18 GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD-----GGGNGCGSGGGGG 70
GD GGDS G + V + GG GGGG GGG GGGG
Sbjct: 457 GDGRPGGDSR*WGPSGVSGKVGSKQSTPPPPHGGFGGGGHSTTTGGGGGSGSGFGGGG 284
Score = 24.3 bits (51), Expect = 9.0
Identities = 11/24 (45%), Positives = 11/24 (45%)
Frame = -2
Query: 8 DGGFADGGSSGDDGCGGDSDGGGG 31
DGG GG G G GGGG
Sbjct: 235 DGGIGGGGDGGGGHSIGLGGGGGG 164
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 53.1 bits (126), Expect(2) = 8e-11
Identities = 32/76 (42%), Positives = 35/76 (45%), Gaps = 6/76 (7%)
Frame = +1
Query: 4 VVVDDGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGG------GGGD 57
+ V++ GG G G GG G GG G GG GGGG GGG
Sbjct: 301 ITVNEAQSRGGGGGGGGGGGGYGGGRGG----------GYGGGGRGGGGGYNRSGGGGGY 450
Query: 58 GGGNGCGSGGGGGCGG 73
GGG G G GGGGG GG
Sbjct: 451 GGGGGYGGGGGGGYGG 498
Score = 51.2 bits (121), Expect = 7e-08
Identities = 34/77 (44%), Positives = 36/77 (46%), Gaps = 7/77 (9%)
Frame = +1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ GG G G G + GGGG G GG GGGGGGG GGG G GGG
Sbjct: 382 GGYGGGGRGG--GGGYNRSGGGG----------GYGGGGGYGGGGGGGYGGGRDRGYGGG 525
Query: 69 -------GGCGGLMWWR 78
GG GG WR
Sbjct: 526 GDRGYSRGGDGGDGGWR 576
Score = 44.7 bits (104), Expect(2) = 7e-08
Identities = 25/64 (39%), Positives = 30/64 (46%), Gaps = 10/64 (15%)
Frame = +1
Query: 20 DGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCG----------SGGGG 69
+G G + G + + + G GG GGG GGG GGG G G SGGGG
Sbjct: 265 EGMNGQNLDGRNITVNEAQSRGGGGGGGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGG 444
Query: 70 GCGG 73
G GG
Sbjct: 445 GYGG 456
Score = 27.7 bits (60), Expect(2) = 8e-11
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +2
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVVVEGD 113
+ V TVV VVTVV +VVV V+ G+
Sbjct: 491 MAVAETVVMVVVVTVVTLVVVTVVMEAGE 577
Score = 26.2 bits (56), Expect(2) = 7e-08
Identities = 15/27 (55%), Positives = 16/27 (58%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
V V + VA VV VVVV VV VVV
Sbjct: 473 VAVAVAMAVAETVVMVVVVTVVTLVVV 553
Score = 24.3 bits (51), Expect(2) = 0.83
Identities = 6/10 (60%), Positives = 7/10 (70%)
Frame = +3
Query: 74 LMWWRRRWWR 83
+ WW RWWR
Sbjct: 387 IWWWWSRWWR 416
Score = 21.9 bits (45), Expect(2) = 0.83
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +2
Query: 84 VVVVTSTVVATAVVTVVV 101
V+VV TVV VVTVV+
Sbjct: 512 VMVVVVTVVTLVVVTVVM 565
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 60.8 bits (146), Expect = 9e-11
Identities = 34/66 (51%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Frame = -3
Query: 9 GGFADGGSSGD-DGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGG 67
GG GGS G G GG GGGG G GG S GGGGGG GGG G G GG
Sbjct: 478 GGSGGGGSQGGGSGSGGGGYGGGG---------SGGSEGGGSSGGGGGGGGGGGGGGYGG 326
Query: 68 GGGCGG 73
GG GG
Sbjct: 325 GGDKGG 308
Score = 58.5 bits (140), Expect = 4e-10
Identities = 32/65 (49%), Positives = 33/65 (50%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG + GG SG G G G GG EG S G GGGGGGG GGG G G G
Sbjct: 466 GGGSQGGGSGSGGGGYGGGGSGGS--------EGGGSSGGGGGGGGGGGGGGYGGGGDKG 311
Query: 69 GGCGG 73
GG GG
Sbjct: 310 GGIGG 296
Score = 54.7 bits (130), Expect(2) = 2e-10
Identities = 30/65 (46%), Positives = 30/65 (46%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
G GGS G G GG GGG G GG S GG GG GGG G GS GG
Sbjct: 625 GSAGGGGSKGGGGYGGGGSQGGG----------GSAGGGGS*GGSGGSAGGGEGSGSQGG 476
Query: 69 GGCGG 73
G GG
Sbjct: 475 GSGGG 461
Score = 53.1 bits (126), Expect(2) = 2e-09
Identities = 34/68 (50%), Positives = 36/68 (52%), Gaps = 3/68 (4%)
Frame = -3
Query: 9 GGFADGGSSGDDGC--GGDSDGGGGVVIEKVVMVEGDCS-GGDSGGGGGGGDGGGNGCGS 65
GG+ GGS G G GG S GG G EG S GG SGGGG G G G+G G
Sbjct: 592 GGYGGGGSQGGGGSAGGGGS*GGSG---GSAGGGEGSGSQGGGSGGGGSQGGGSGSGGGG 422
Query: 66 GGGGGCGG 73
GGGG GG
Sbjct: 421 YGGGGSGG 398
Score = 50.8 bits (120), Expect = 9e-08
Identities = 31/64 (48%), Positives = 33/64 (51%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG+ GGS G +G GG S GGGG GG GGGGGGG GGG G G G
Sbjct: 427 GGYGGGGSGGSEG-GGSSGGGGG--------------GG--GGGGGGGYGGGGDKGGGIG 299
Query: 69 GGCG 72
G G
Sbjct: 298 GDKG 287
Score = 47.4 bits (111), Expect = 1e-06
Identities = 31/71 (43%), Positives = 31/71 (43%), Gaps = 6/71 (8%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG------GGNG 62
GG GGSSG G GG GGGG GGD GGG GG G GG G
Sbjct: 403 GGSEGGGSSGGGGGGGGGGGGGGYG-----------GGGDKGGGIGGDKGDYHHGKGGKG 257
Query: 63 CGSGGGGGCGG 73
GG G GG
Sbjct: 256 DKDGGDKGGGG 224
Score = 47.0 bits (110), Expect = 1e-06
Identities = 28/67 (41%), Positives = 30/67 (43%), Gaps = 2/67 (2%)
Frame = -3
Query: 9 GGFADGGSS--GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSG 66
GG A GG S G G G +G G +G SGG GGG G GGG G G
Sbjct: 559 GGSAGGGGS*GGSGGSAGGGEGSGS---------QGGGSGGGGSQGGGSGSGGGGYGGGG 407
Query: 67 GGGGCGG 73
GG GG
Sbjct: 406 SGGSEGG 386
Score = 41.6 bits (96), Expect = 5e-05
Identities = 23/51 (45%), Positives = 26/51 (50%), Gaps = 5/51 (9%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMV-----EGDCSGGDSGGGGG 54
GG GG G G GG D GGG+ +K +GD GGD GGGGG
Sbjct: 373 GGGGGGGGGGGGGYGGGGDKGGGIGGDKGDYHHGKGGKGDKDGGDKGGGGG 221
Score = 40.0 bits (92), Expect = 2e-04
Identities = 26/65 (40%), Positives = 30/65 (46%), Gaps = 2/65 (3%)
Frame = -3
Query: 8 DGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGG--GGGGGDGGGNGCGS 65
+GG + GG G G GG GGG +G GGD G G GG G +G
Sbjct: 394 EGGGSSGGGGGGGGGGGGGGYGGG-------GDKGGGIGGDKGDYHHGKGGKGDKDGGDK 236
Query: 66 GGGGG 70
GGGGG
Sbjct: 235 GGGGG 221
Score = 38.9 bits (89), Expect = 4e-04
Identities = 17/28 (60%), Positives = 18/28 (63%)
Frame = -3
Query: 46 GGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
GG +GGGG G GG G GS GGGG G
Sbjct: 628 GGSAGGGGSKGGGGYGGGGSQGGGGSAG 545
Score = 38.5 bits (88), Expect(2) = 9e-05
Identities = 21/48 (43%), Positives = 22/48 (45%)
Frame = -3
Query: 26 SDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
S+GGGG S G G GGGG GGG G GG G GG
Sbjct: 640 SEGGGG-------------SAGGGGSKGGGGYGGGGSQGGGGSAGGGG 536
Score = 25.0 bits (53), Expect = 5.3
Identities = 11/25 (44%), Positives = 12/25 (48%)
Frame = -3
Query: 7 DDGGFADGGSSGDDGCGGDSDGGGG 31
D G + G D GGD GGGG
Sbjct: 295 DKGDYHHGKGGKGDKDGGDKGGGGG 221
Score = 25.0 bits (53), Expect(2) = 2e-10
Identities = 15/27 (55%), Positives = 16/27 (58%)
Frame = -1
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VVVV V+A VV V V VEA VV
Sbjct: 405 VVVVKEEVLAVVVVEVEAEVEVEATVV 325
Score = 23.1 bits (48), Expect(2) = 2e-09
Identities = 14/26 (53%), Positives = 14/26 (53%)
Frame = -1
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVV 109
V VV VV V VVVV VEA V
Sbjct: 423 VTVVVEVVVVKEEVLAVVVVEVEAEV 346
Score = 21.6 bits (44), Expect(2) = 9e-05
Identities = 14/27 (51%), Positives = 15/27 (54%)
Frame = -1
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVVVE 111
VVV VV V+ VVVV V V VE
Sbjct: 417 VVVEVVVVKEEVLAVVVVEVEAEVEVE 337
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 53.9 bits (128), Expect(2) = 2e-10
Identities = 37/81 (45%), Positives = 39/81 (47%), Gaps = 16/81 (19%)
Frame = -1
Query: 9 GGFADGGSSG---DDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD---GGGNG 62
GG DGG G GGD DGGG E G+CS GGGGGGGD GGG+G
Sbjct: 372 GGGGDGGGGELY*IGGGGGDGDGGGW*GAECGDGGGGECSSQGGGGGGGGGDSIQGGGDG 193
Query: 63 CGSGG----------GGGCGG 73
G GG GGG GG
Sbjct: 192 GGGGGE*TGGGGE*TGGGGGG 130
Score = 49.3 bits (116), Expect = 3e-07
Identities = 34/84 (40%), Positives = 36/84 (42%), Gaps = 18/84 (21%)
Frame = -1
Query: 8 DGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSG-----------GGGGGG 56
DGG G GD G G S GGG ++G GG G GGGGGG
Sbjct: 309 DGGGW*GAECGDGGGGECSSQGGGGGGGGGDSIQGGGDGGGGGGE*TGGGGE*TGGGGGG 130
Query: 57 D-------GGGNGCGSGGGGGCGG 73
D GGG CG GG GG GG
Sbjct: 129 DK*G*TGGGGGGECGGGGDGGGGG 58
Score = 48.9 bits (115), Expect = 3e-07
Identities = 32/69 (46%), Positives = 35/69 (50%), Gaps = 5/69 (7%)
Frame = -1
Query: 10 GFADGGSSGDD--GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGG 67
G DGG G + G GG+ GGGG GD G GGGGG GGG+G G GG
Sbjct: 207 GGGDGGGGGGE*TGGGGE*TGGGG---------GGDK*G*TGGGGGGECGGGGDGGGGGG 55
Query: 68 ---GGGCGG 73
GGG GG
Sbjct: 54 EYVGGGAGG 28
Score = 47.0 bits (110), Expect = 1e-06
Identities = 28/65 (43%), Positives = 29/65 (44%)
Frame = -1
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G D G GGGG +C GG GGGGGG GG G G
Sbjct: 162 GGE*TGGGGGGDK*G*TGGGGGG-----------ECGGGGDGGGGGGEYVGGGAGGEYTG 16
Query: 69 GGCGG 73
GG GG
Sbjct: 15 GGGGG 1
Score = 40.8 bits (94), Expect = 9e-05
Identities = 22/50 (44%), Positives = 26/50 (52%)
Frame = -1
Query: 7 DDGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGG 56
D G GG G+ G GGD GGGG + G +GG+ GGGGGG
Sbjct: 129 DK*G*TGGGGGGECGGGGDGGGGGGEYV-------GGGAGGEYTGGGGGG 1
Score = 34.3 bits (77), Expect = 0.009
Identities = 23/57 (40%), Positives = 25/57 (43%)
Frame = -1
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGGLMWW 77
G GG DGGGG + +G GGGG GGG GGGGG G W
Sbjct: 429 GAGGGDDGGGGEL***------------TGVGGGGDGGGGELY*IGGGGGDGDGGGW 295
Score = 27.3 bits (59), Expect = 1.1
Identities = 8/12 (66%), Positives = 10/12 (82%)
Frame = -3
Query: 76 WWRRRWWRVVVV 87
WWR RWWR +V+
Sbjct: 421 WWR*RWWR*IVM 386
Score = 25.4 bits (54), Expect(2) = 2e-10
Identities = 8/14 (57%), Positives = 10/14 (71%), Gaps = 3/14 (21%)
Frame = -3
Query: 74 LMWWRRR---WWRV 84
+ WWRRR WWR+
Sbjct: 91 MWWWRRRWRWWWRI 50
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 54.3 bits (129), Expect(2) = 4e-10
Identities = 34/68 (50%), Positives = 35/68 (51%), Gaps = 3/68 (4%)
Frame = -2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCS---GGDSGGGGGGGDGGGNGCGS 65
GG DGG GCG DGGGG +CS GGD GGGGG GGG G GS
Sbjct: 521 GGDGDGGGW*GAGCG---DGGGG-----------ECSSHGGGDGGGGGGSIQGGGGGGGS 384
Query: 66 GGGGGCGG 73
GGG GG
Sbjct: 383 IQGGGDGG 360
Score = 53.5 bits (127), Expect = 1e-08
Identities = 28/62 (45%), Positives = 34/62 (54%)
Frame = -2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG + G G G GG +GGGG + + V G+ GG+ GGGGG GGG G G G
Sbjct: 179 GGGENTGGGGGGGDGGGGEGGGGDLTQYV----GEGGGGEGGGGGGE*TGGGGEGGGGDG 12
Query: 69 GG 70
GG
Sbjct: 11 GG 6
Score = 52.8 bits (125), Expect = 2e-08
Identities = 32/72 (44%), Positives = 36/72 (49%), Gaps = 7/72 (9%)
Frame = -2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGG-----GGGGDGGGNG- 62
GG GG G G DGGGG + G+ +GG GG GGGG+GGG G
Sbjct: 422 GGSIQGGGGGGGSIQGGGDGGGGG--GE*TGGGGE*TGGGGGGDK*G*TGGGGEGGGGGG 249
Query: 63 -CGSGGGGGCGG 73
CG GGGG C G
Sbjct: 248 ECGGGGGGECCG 213
Score = 51.6 bits (122), Expect = 5e-08
Identities = 34/76 (44%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Frame = -2
Query: 8 DGGFADGGSSGDDG------CGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD---- 57
DGG G G G GGD GGGG E GG+ GGGGGGD
Sbjct: 437 DGGGGGGSIQGGGGGGGSIQGGGDGGGGGG---------E*TGGGGE*TGGGGGGDK*G* 285
Query: 58 GGGNGCGSGGGGGCGG 73
GG G G GGGG CGG
Sbjct: 284 TGGGGEGGGGGGECGG 237
Score = 48.9 bits (115), Expect(2) = 9e-08
Identities = 31/71 (43%), Positives = 33/71 (45%), Gaps = 5/71 (7%)
Frame = -2
Query: 9 GGFADGGSSG---DDGCGGDSDGGG--GVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGC 63
GG DGG G GGD DGGG G GD GG+ GGG GGG G
Sbjct: 572 GGGGDGGGGELY*TGGGGGDGDGGGW*GAGC-------GDGGGGECSSHGGGDGGGGGGS 414
Query: 64 GSGGGGGCGGL 74
GGGGG G +
Sbjct: 413 IQGGGGGGGSI 381
Score = 48.5 bits (114), Expect = 4e-07
Identities = 36/84 (42%), Positives = 38/84 (44%), Gaps = 19/84 (22%)
Frame = -2
Query: 9 GGFADGGSSGDDGCGGDSD------GGGGVVIEKVVMVEGDCSGGDSGGG-GGGGD---- 57
GG GG G + CGG + GGGG G GGD GGG GGGGD
Sbjct: 257 GGGECGGGGGGECCGGGGEL**YCTGGGGE------NTGGGGGGGDGGGGEGGGGDLTQY 96
Query: 58 ----GGGNGCGSG----GGGGCGG 73
GGG G G G GGGG GG
Sbjct: 95 VGEGGGGEGGGGGGE*TGGGGEGG 24
Score = 48.5 bits (114), Expect = 4e-07
Identities = 31/73 (42%), Positives = 33/73 (44%), Gaps = 6/73 (8%)
Frame = -2
Query: 7 DDGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSG------GGGGGGDGGG 60
D G GG G G GG+ GGGG G+C GG GGGG GGG
Sbjct: 299 DK*G*TGGGGEGGGG-GGECGGGGG----------GECCGGGGEL**YCTGGGGENTGGG 153
Query: 61 NGCGSGGGGGCGG 73
G G GGGG GG
Sbjct: 152 GGGGDGGGGEGGG 114
Score = 46.2 bits (108), Expect = 2e-06
Identities = 27/64 (42%), Positives = 32/64 (49%), Gaps = 3/64 (4%)
Frame = -2
Query: 9 GGFADGGSSGDD---GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGS 65
GG +GG G + G GG+ GGGG + C+GG GGGG GG G G
Sbjct: 281 GGGGEGGGGGGECGGGGGGECCGGGGEL**Y-------CTGGGGENTGGGGGGGDGGGGE 123
Query: 66 GGGG 69
GGGG
Sbjct: 122 GGGG 111
Score = 45.8 bits (107), Expect = 3e-06
Identities = 35/87 (40%), Positives = 37/87 (42%), Gaps = 23/87 (26%)
Frame = -2
Query: 10 GFADGGSSGDD--GCGGDSDGGGGVVIEKVVMVEGD----CSGGDSGGGGGG---GDGGG 60
G DGG G + G GG+ GGGG GD GG GGGGGG G GGG
Sbjct: 377 GGGDGGGGGGE*TGGGGE*TGGGG---------GGDK*G*TGGGGEGGGGGGECGGGGGG 225
Query: 61 NGCGSGG--------------GGGCGG 73
CG GG GGG GG
Sbjct: 224 ECCGGGGEL**YCTGGGGENTGGGGGG 144
Score = 38.5 bits (88), Expect = 5e-04
Identities = 26/59 (44%), Positives = 31/59 (52%), Gaps = 6/59 (10%)
Frame = -2
Query: 8 DGGFADGGSSGD------DGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG 60
DGG +GG GD +G GG+ GGG G+ +GG GG GGGGDGGG
Sbjct: 140 DGGGGEGGG-GDLTQYVGEGGGGEGGGGG-----------GE*TGG--GGEGGGGDGGG 6
Score = 35.4 bits (80), Expect = 0.004
Identities = 23/57 (40%), Positives = 27/57 (47%)
Frame = -2
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGGLMWW 77
G GG +DGGGG + +G GGGG GGG +GGGGG G W
Sbjct: 629 GVGGGNDGGGGEL***------------TGVGGGGDGGGGELY*TGGGGGDGDGGGW 495
Score = 30.4 bits (67), Expect(2) = 0.002
Identities = 20/51 (39%), Positives = 21/51 (40%), Gaps = 4/51 (7%)
Frame = -2
Query: 28 GGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG----NGCGSGGGGGCGGL 74
GGGG + M G GGG G GG G G GG GG G L
Sbjct: 692 GGGGGDAYETPMTGGKGPSYTGVGGGNDGGGGEL***TGVGGGGDGGGGEL 540
Score = 30.4 bits (67), Expect = 0.12
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = -2
Query: 6 VDDGGFADGGSSGDDGCGGDSDGGGG 31
V +GG +GG G + GG +GGGG
Sbjct: 95 VGEGGGGEGGGGGGE*TGGGGEGGGG 18
Score = 25.0 bits (53), Expect(2) = 0.002
Identities = 7/10 (70%), Positives = 8/10 (80%)
Frame = -1
Query: 76 WWRRRWWRVV 85
WWR RWW +V
Sbjct: 522 WWRWRWWWMV 493
Score = 24.3 bits (51), Expect(2) = 4e-10
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = -1
Query: 76 WWRRRWWR 83
WW R WWR
Sbjct: 258 WWGRMWWR 235
Score = 21.6 bits (44), Expect(2) = 9e-08
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = -1
Query: 76 WWRRRWW 82
WWR WW
Sbjct: 375 WWRWGWW 355
>TC207679 similar to UP|Q7XJI7 (Q7XJI7) Glycine-rich protein TomR2, partial
(22%)
Length = 425
Score = 58.5 bits (140), Expect = 4e-10
Identities = 32/68 (47%), Positives = 37/68 (54%), Gaps = 2/68 (2%)
Frame = +3
Query: 8 DGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGG 67
+GG+ GG SG GG + GGGG G GG +G GGGG GGG G G+GG
Sbjct: 21 EGGYGSGGGSGGGYGGGAAGGGGG----------GSGGGGGAGSGGGGAHGGGYGGGAGG 170
Query: 68 --GGGCGG 73
GGG GG
Sbjct: 171GEGGGHGG 194
Score = 53.9 bits (128), Expect(2) = 5e-10
Identities = 29/62 (46%), Positives = 30/62 (47%)
Frame = +3
Query: 12 ADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGGGC 71
A G G G GG S GG GG + GGGGGG GGG G GSGGGG
Sbjct: 6 AGGAHEGGYGSGGGSGGG---------------YGGGAAGGGGGGSGGGGGAGSGGGGAH 140
Query: 72 GG 73
GG
Sbjct: 141GG 146
Score = 31.2 bits (69), Expect = 0.073
Identities = 14/28 (50%), Positives = 15/28 (53%)
Frame = +3
Query: 46 GGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G GG G GGG+G G GGG GG
Sbjct: 3 GAGGAHEGGYGSGGGSGGGYGGGAAGGG 86
Score = 24.3 bits (51), Expect(2) = 5e-10
Identities = 6/7 (85%), Positives = 6/7 (85%)
Frame = +2
Query: 76 WWRRRWW 82
WW RRWW
Sbjct: 164 WWWRRWW 184
>TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, complete
Length = 1937
Score = 50.4 bits (119), Expect(2) = 8e-10
Identities = 31/73 (42%), Positives = 35/73 (47%), Gaps = 4/73 (5%)
Frame = +2
Query: 4 VVVDDGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD----GG 59
+ V++ GG G G GG + GGGG GG SGGGGGGG G
Sbjct: 1454 ITVNEAQSRGGGGGGGGGGGGYNRGGGGY-------------GGRSGGGGGGGGYRSRDG 1594
Query: 60 GNGCGSGGGGGCG 72
G G G GGGGG G
Sbjct: 1595 GYGGGYGGGGGGG 1633
Score = 48.5 bits (114), Expect = 4e-07
Identities = 33/72 (45%), Positives = 34/72 (46%), Gaps = 2/72 (2%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG--NGCGSG 66
GG G + G G GG S GGGG GG GGGGGGG GGG G G
Sbjct: 1499 GGGGGGYNRGGGGYGGRSGGGGG---GGGYRSRDGGYGGGYGGGGGGGYGGGRDRGYSRG 1669
Query: 67 GGGGCGGLMWWR 78
G GG GG WR
Sbjct: 1670 GDGGDGG---WR 1696
Score = 43.5 bits (101), Expect(2) = 2e-07
Identities = 22/41 (53%), Positives = 24/41 (57%), Gaps = 4/41 (9%)
Frame = +2
Query: 37 VVMVEGDCSGGDSGGGGGGGD----GGGNGCGSGGGGGCGG 73
+ + E GG GGGGGGG GGG G SGGGGG GG
Sbjct: 1454 ITVNEAQSRGGGGGGGGGGGGYNRGGGGYGGRSGGGGGGGG 1576
Score = 26.9 bits (58), Expect(2) = 8e-10
Identities = 15/31 (48%), Positives = 18/31 (57%)
Frame = +3
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVVEGDC 114
VVVV V T V VVV+VV+EA + C
Sbjct: 1620 VVVVVMAVAETVVTLVVVMVVMEAGETKA*C 1712
Score = 25.8 bits (55), Expect(2) = 2e-07
Identities = 14/22 (63%), Positives = 16/22 (72%)
Frame = +3
Query: 84 VVVVTSTVVATAVVTVVVVVVV 105
VVVV VA VVT+VVV+VV
Sbjct: 1617 VVVVVVMAVAETVVTLVVVMVV 1682
Score = 25.4 bits (54), Expect(2) = 0.60
Identities = 14/27 (51%), Positives = 16/27 (58%)
Frame = +3
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VV V VV T VTVV+ V + VVV
Sbjct: 1548 VVAVAVAVVDTVAVTVVMAVAMVVVVV 1628
Score = 21.2 bits (43), Expect(2) = 0.60
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = +1
Query: 76 WWRRRWW 82
WWR WW
Sbjct: 1525 WWRWIWW 1545
>TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (11%)
Length = 678
Score = 56.6 bits (135), Expect = 2e-09
Identities = 31/67 (46%), Positives = 35/67 (51%), Gaps = 1/67 (1%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGG-GDGGGNGCGSGG 67
GG G GD G GG DGG G ++ + G GGD G GGGG G GG G G G
Sbjct: 382 GGDGRFGGEGDGGKGGGGDGGKGGGVDSGIG-GGKGGGGDGGNGGGGDGGNGGGGDGGNG 206
Query: 68 GGGCGGL 74
GGG G+
Sbjct: 205 GGGASGI 185
Score = 56.2 bits (134), Expect = 2e-09
Identities = 31/66 (46%), Positives = 33/66 (49%), Gaps = 2/66 (3%)
Frame = -3
Query: 9 GGFADGGSSG--DDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSG 66
GG DGG G D G GG GGG +G GG GG GGGGDGG G G+
Sbjct: 340 GGGGDGGKGGGVDSGIGGGKGGGG----------DGGNGGGGDGGNGGGGDGGNGGGGAS 191
Query: 67 GGGGCG 72
G GG G
Sbjct: 190 GIGGGG 173
Score = 53.9 bits (128), Expect = 1e-08
Identities = 32/87 (36%), Positives = 38/87 (42%), Gaps = 20/87 (22%)
Frame = -3
Query: 6 VDDGGFADGGSS--------------------GDDGCGGDSDGGGGVVIEKVVMVEGDCS 45
+ +GG GG + GD G GG DGG G ++ + G
Sbjct: 451 IGNGGSGGGGGASGGNGGGGASGGGDGRFGGEGDGGKGGGGDGGKGGGVDS--GIGGGKG 278
Query: 46 GGDSGGGGGGGDGGGNGCGSGGGGGCG 72
GG GG GGGGDGG G G GG GG G
Sbjct: 277 GGGDGGNGGGGDGGNGGGGDGGNGGGG 197
Score = 53.5 bits (127), Expect = 1e-08
Identities = 38/101 (37%), Positives = 43/101 (41%), Gaps = 1/101 (0%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG-GGNGCGSGG 67
GG D G G G GGD GGG G+ GGD G GGGG G GG G GG
Sbjct: 316 GGGVDSGIGGGKGGGGDGGNGGGGD-------GGNGGGGDGGNGGGGASGIGGGGHSLGG 158
Query: 68 GGGCGGLMWWRRRWWRVVVVTSTVVATAVVTVVVVVVVEAV 108
GGG + ++ T + T V V V V E V
Sbjct: 157 GGGSSHSL-------HIMCGTWSSFPTVKVVWVAVAVTETV 56
Score = 49.7 bits (117), Expect(2) = 9e-09
Identities = 27/62 (43%), Positives = 29/62 (46%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG +GGS G G G + GGG SGG G GG GDGG G G GG
Sbjct: 457 GGIGNGGSGGGGGASGGNGGGGA-------------SGGGDGRFGGEGDGGKGGGGDGGK 317
Query: 69 GG 70
GG
Sbjct: 316 GG 311
Score = 45.1 bits (105), Expect = 5e-06
Identities = 29/62 (46%), Positives = 30/62 (47%), Gaps = 3/62 (4%)
Frame = -3
Query: 15 GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGG-GGGDG--GGNGCGSGGGGGC 71
G G G GG GGG SGG+ GGG GGGDG GG G G GGGG
Sbjct: 466 GPFGGIGNGGSGGGGGA-------------SGGNGGGGASGGGDGRFGGEGDGGKGGGGD 326
Query: 72 GG 73
GG
Sbjct: 325 GG 320
Score = 40.8 bits (94), Expect = 9e-05
Identities = 33/100 (33%), Positives = 36/100 (36%), Gaps = 9/100 (9%)
Frame = -3
Query: 8 DGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGG 67
DGG G GD G GG DGG +GGGG G GGG GG
Sbjct: 268 DGGNGGG---GDGGNGGGGDGG-------------------NGGGGASGIGGGGHSLGGG 155
Query: 68 GGGCGGLMWWRRRW---------WRVVVVTSTVVATAVVT 98
GG L W W V VT TV + +T
Sbjct: 154 GGSSHSLHIMCGTWSSFPTVKVVWVAVAVTETVEDRSKIT 35
Score = 37.0 bits (84), Expect = 0.001
Identities = 18/31 (58%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Frame = -3
Query: 45 SGGDSGGGGGGGDGGGNGC--GSGGGGGCGG 73
S G GG G GG GGG G G+GGGG GG
Sbjct: 472 SPGPFGGIGNGGSGGGGGASGGNGGGGASGG 380
Score = 28.1 bits (61), Expect(2) = 0.053
Identities = 13/27 (48%), Positives = 14/27 (51%)
Frame = -3
Query: 47 GDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G GGG G G G+GG GG GG
Sbjct: 499 GSRGGGTRYSPGPFGGIGNGGSGGGGG 419
Score = 25.4 bits (54), Expect = 4.0
Identities = 15/28 (53%), Positives = 17/28 (60%)
Frame = -1
Query: 83 RVVVVTSTVVATAVVTVVVVVVVEAVVV 110
R+V V VV VVV+VVVE VVV
Sbjct: 462 RLVGSAMGVAVVVVVQVVVMVVVEPVVV 379
Score = 24.3 bits (51), Expect(2) = 9e-09
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = -1
Query: 86 VVTSTVVATAVVTVVVVVVVEAVVV 110
V+ TVV VVTVV V+VV V V
Sbjct: 270 VMVVTVVEVMVVTVVEVMVVTVVEV 196
Score = 22.3 bits (46), Expect(2) = 0.053
Identities = 13/27 (48%), Positives = 15/27 (55%)
Frame = -1
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVVVE 111
VVV VV VV VVVV+V+ E
Sbjct: 432 VVVVVQVVVMVVVEPVVVVMVDLAAKE 352
>TC230423 similar to UP|Q41187 (Q41187) Glycine-rich protein (Fragment),
partial (40%)
Length = 522
Score = 55.5 bits (132), Expect = 4e-09
Identities = 31/65 (47%), Positives = 34/65 (51%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG+ G G GG GG G G GG +GGG GGG GGG G G+GGG
Sbjct: 83 GGGIGGGAGGGGGAGGGFGGGAGGGA-------GGGFGGGAGGGAGGGFGGGAGGGAGGG 241
Query: 69 GGCGG 73
G GG
Sbjct: 242 FGGGG 256
Score = 54.7 bits (130), Expect = 6e-09
Identities = 30/62 (48%), Positives = 33/62 (52%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG A GG G GG + GG G G +GG +GGG GGG GGG G G GGG
Sbjct: 95 GGGAGGGGGAGGGFGGGAGGGAGGGF-------GGGAGGGAGGGFGGGAGGGAGGGFGGG 253
Query: 69 GG 70
GG
Sbjct: 254 GG 259
Score = 52.8 bits (125), Expect(2) = 2e-09
Identities = 30/66 (45%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Frame = +2
Query: 9 GGFADGGSSGDDGCGGDSDGG-GGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGG 67
GG GG G G GG GG GG V + G G G GGG G G G G G G
Sbjct: 5 GGGVGGGIGGGGGAGGGFGGGHGGGVGGGIGGGAGGGGGAGGGFGGGAGGGAGGGFGGGA 184
Query: 68 GGGCGG 73
GGG GG
Sbjct: 185GGGAGG 202
Score = 41.2 bits (95), Expect(3) = 1e-06
Identities = 19/33 (57%), Positives = 20/33 (60%)
Frame = +2
Query: 41 EGDCSGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
+G GG GGGGG G G G G G G GGG GG
Sbjct: 2 KGGGVGGGIGGGGGAGGGFGGGHGGGVGGGIGG 100
Score = 26.6 bits (57), Expect = 1.8
Identities = 7/9 (77%), Positives = 8/9 (88%)
Frame = +1
Query: 76 WWRRRWWRV 84
WW RRWWR+
Sbjct: 31 WWWRRWWRL 57
Score = 23.1 bits (48), Expect(2) = 2e-09
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = +1
Query: 76 WWRRRWWR 83
WW RWWR
Sbjct: 232 WWWFRWWR 255
Score = 22.7 bits (47), Expect(3) = 1e-06
Identities = 14/25 (56%), Positives = 15/25 (60%)
Frame = +3
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAV 108
VV V VVA VV V V+VVV V
Sbjct: 177 VVQVEELVVALEVVQVGVLVVVSVV 251
Score = 21.6 bits (44), Expect(3) = 1e-06
Identities = 7/11 (63%), Positives = 7/11 (63%)
Frame = +1
Query: 73 GLMWWRRRWWR 83
G WWR RW R
Sbjct: 85 GRNWWRCRWRR 117
>TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%)
Length = 1030
Score = 55.5 bits (132), Expect = 4e-09
Identities = 32/68 (47%), Positives = 37/68 (54%)
Frame = -1
Query: 10 GFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGG 69
G GG +G G GGD GGGG ++EG G +GGGGGGG +G G GGGG
Sbjct: 313 GGGGGGETGGGGGGGDKGGGGGGGWLLY*LLEG*YIG--AGGGGGGGWYEASGGGGGGGG 140
Query: 70 GCGGLMWW 77
G G WW
Sbjct: 139 GYG**GWW 116
Score = 42.4 bits (98), Expect(2) = 5e-07
Identities = 22/47 (46%), Positives = 25/47 (52%), Gaps = 14/47 (29%)
Frame = -1
Query: 40 VEGDCSGGDSGGGGGGGDGGGNG--------------CGSGGGGGCG 72
V G GG++GGGGGGGD GG G G+GGGGG G
Sbjct: 319 VTGGGGGGETGGGGGGGDKGGGGGGGWLLY*LLEG*YIGAGGGGGGG 179
Score = 40.0 bits (92), Expect = 2e-04
Identities = 17/25 (68%), Positives = 18/25 (72%)
Frame = -1
Query: 49 SGGGGGGGDGGGNGCGSGGGGGCGG 73
+GGGGGG GGG G G GGGG GG
Sbjct: 316 TGGGGGGETGGGGGGGDKGGGGGGG 242
Score = 25.4 bits (54), Expect(2) = 5e-07
Identities = 8/32 (25%), Positives = 19/32 (59%)
Frame = -3
Query: 74 LMWWRRRWWRVVVVTSTVVATAVVTVVVVVVV 105
L W RRRWW + +V + + +++++++
Sbjct: 167 LRWRRRRWWWIWIVRVVGIWRCIWNMMLLILM 72
>TC233801 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (5%)
Length = 394
Score = 54.7 bits (130), Expect = 6e-09
Identities = 40/100 (40%), Positives = 43/100 (43%)
Frame = -2
Query: 10 GFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGG 69
GF GG G GGD+ GGG GD G GGGG GGG+ G GGGG
Sbjct: 393 GFGGGGHLIITGGGGDAGNGGG---------------GDGGIGGGGDGGGGHSFGCGGGG 259
Query: 70 GCGGLMWWRRRWWRVVVVTSTVVATAVVTVVVVVVVEAVV 109
G + R W T V V VVVV EAVV
Sbjct: 258 GSSHSLPIMRGTWS--------FPTVEVVVWVVVVDEAVV 163
Score = 25.0 bits (53), Expect = 5.3
Identities = 12/22 (54%), Positives = 12/22 (54%)
Frame = -2
Query: 9 GGFADGGSSGDDGCGGDSDGGG 30
GG DGG GCGG GGG
Sbjct: 312 GGGGDGGGGHSFGCGG---GGG 256
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 53.9 bits (128), Expect = 1e-08
Identities = 35/77 (45%), Positives = 38/77 (48%), Gaps = 12/77 (15%)
Frame = +3
Query: 9 GGFADGGSSGDDGC--GGDSDGGGGVVIE--KVVMVEGDCSGGDSG------GGGG--GG 56
GG+ GG G G GG GGGG + + DCSGG G GGGG GG
Sbjct: 357 GGYGGGGGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGG 536
Query: 57 DGGGNGCGSGGGGGCGG 73
GGG G GGGGG GG
Sbjct: 537 GGGGRYVGGGGGGGGGG 587
Score = 51.2 bits (121), Expect = 7e-08
Identities = 33/76 (43%), Positives = 33/76 (43%), Gaps = 4/76 (5%)
Frame = +3
Query: 2 VAVVVDDGGFADGGSSGDDGC----GGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD 57
V V DG G G DG GG GGGG GG G GGGGG
Sbjct: 243 VDVTGPDGASVQGTRRGGDGGRSYGGGRGGGGGGY-------------GGGGGYGGGGGY 383
Query: 58 GGGNGCGSGGGGGCGG 73
GGG G G GG GG GG
Sbjct: 384 GGGGGYGGGGRGGGGG 431
Score = 41.6 bits (96), Expect = 5e-05
Identities = 32/77 (41%), Positives = 33/77 (42%), Gaps = 14/77 (18%)
Frame = +3
Query: 8 DGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNG----- 62
DGG + GG G G GG GGGG G G GGG GGG GG G
Sbjct: 297 DGGRSYGGGRG--GGGGGYGGGGGY---------GGGGGYGGGGGYGGGGRGGGGGACYN 443
Query: 63 CG---------SGGGGG 70
CG SGGGGG
Sbjct: 444 CGESGHLARDCSGGGGG 494
Score = 25.4 bits (54), Expect = 4.0
Identities = 17/26 (65%), Positives = 18/26 (68%), Gaps = 2/26 (7%)
Frame = +1
Query: 86 VVTSTVVAT--AVVTVVVVVVVEAVV 109
V T VVAT AVVTVVV VEAV+
Sbjct: 358 VATVVVVATVEAVVTVVVEEAVEAVL 435
Score = 25.0 bits (53), Expect = 5.3
Identities = 13/23 (56%), Positives = 13/23 (56%)
Frame = +1
Query: 87 VTSTVVATAVVTVVVVVVVEAVV 109
V AV TVVVV VEAVV
Sbjct: 331 VVEVATVEAVATVVVVATVEAVV 399
Score = 24.3 bits (51), Expect = 9.0
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = +1
Query: 85 VVVTSTVVATAVVTVVVVVVVEAVV 109
V +TVV A V VV VVVE V
Sbjct: 349 VEAVATVVVVATVEAVVTVVVEEAV 423
>TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, complete
Length = 1145
Score = 48.5 bits (114), Expect = 4e-07
Identities = 33/72 (45%), Positives = 34/72 (46%), Gaps = 2/72 (2%)
Frame = -3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGG--NGCGSG 66
GG G + G G GG S GGGG GG GGGGGGG GGG G G
Sbjct: 378 GGGGGGYNRGGGGYGGRSGGGGG---GGGYRSRDGGYGGGYGGGGGGGYGGGRDRGYSRG 208
Query: 67 GGGGCGGLMWWR 78
G GG GG WR
Sbjct: 207 GDGGDGG---WR 181
Score = 46.6 bits (109), Expect(2) = 1e-08
Identities = 29/62 (46%), Positives = 30/62 (47%), Gaps = 4/62 (6%)
Frame = -3
Query: 15 GSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD----GGGNGCGSGGGGG 70
G G G GG + GGGG GG SGGGGGGG GG G G GGGGG
Sbjct: 390 GGGGGGGGGGYNRGGGGY-------------GGRSGGGGGGGGYRSRDGGYGGGYGGGGG 250
Query: 71 CG 72
G
Sbjct: 249 GG 244
Score = 42.7 bits (99), Expect(2) = 3e-07
Identities = 21/39 (53%), Positives = 24/39 (60%), Gaps = 2/39 (5%)
Frame = -3
Query: 37 VVMVEGDCSGGDSGGGGGGGDGGGNGCG--SGGGGGCGG 73
+ + E GG GGGGGG + GG G G SGGGGG GG
Sbjct: 417 ITVNEAQSRGGGGGGGGGGYNRGGGGYGGRSGGGGGGGG 301
Score = 26.9 bits (58), Expect(2) = 1e-08
Identities = 15/31 (48%), Positives = 18/31 (57%)
Frame = -1
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVVEGDC 114
VVVV V T V VVV+VV+EA + C
Sbjct: 257 VVVVVMAVAETVVTLVVVMVVMEAGETKA*C 165
Score = 25.8 bits (55), Expect(2) = 3e-07
Identities = 14/22 (63%), Positives = 16/22 (72%)
Frame = -1
Query: 84 VVVVTSTVVATAVVTVVVVVVV 105
VVVV VA VVT+VVV+VV
Sbjct: 260 VVVVVVMAVAETVVTLVVVMVV 195
Score = 25.4 bits (54), Expect(2) = 0.63
Identities = 14/27 (51%), Positives = 16/27 (58%)
Frame = -1
Query: 84 VVVVTSTVVATAVVTVVVVVVVEAVVV 110
VV V VV T VTVV+ V + VVV
Sbjct: 329 VVAVAVAVVDTVAVTVVMAVAMVVVVV 249
Score = 21.2 bits (43), Expect(2) = 0.63
Identities = 5/7 (71%), Positives = 5/7 (71%)
Frame = -2
Query: 76 WWRRRWW 82
WWR WW
Sbjct: 352 WWRWIWW 332
>TC224734 homologue to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (48%)
Length = 990
Score = 53.1 bits (126), Expect = 2e-08
Identities = 39/80 (48%), Positives = 41/80 (50%), Gaps = 15/80 (18%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDG---- 58
GG DG G D G GGD DGGGG + +GD GGD GGGG GDG
Sbjct: 300 GGGGDGDGGGGDL***GGGGDGDGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGD 121
Query: 59 ----GGNGCGSGGGG-GCGG 73
GG G GSGGGG G GG
Sbjct: 120 L***GGGGDGSGGGGDGEGG 61
Score = 52.8 bits (125), Expect = 2e-08
Identities = 34/66 (51%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG-GGNGC 63
GG DG G D G GGD DGGGG + +GD GGD GGGGDG GG G
Sbjct: 252 GGGGDGDGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGSGGGGD 73
Query: 64 GSGGGG 69
G GGGG
Sbjct: 72 GEGGGG 55
Score = 49.7 bits (117), Expect = 2e-07
Identities = 33/76 (43%), Positives = 34/76 (44%), Gaps = 14/76 (18%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDC----SGGDSGGGGGGGDGGG 60
GG DG G D G GGD DGGGG D GGD GGGG G+GGG
Sbjct: 204 GGGGDGDGGGGDL***GGGGDGDGGGG-----------DL***GGGGDGSGGGGDGEGGG 58
Query: 61 ------NGCGSGGGGG 70
G G G GGG
Sbjct: 57 GDL**YGGGGEGDGGG 10
Score = 42.7 bits (99), Expect(2) = 2e-06
Identities = 28/58 (48%), Positives = 31/58 (53%), Gaps = 8/58 (13%)
Frame = -1
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGG------NGCGSGGGGG 70
G GG+ DGGGG + +GD GGD GGGG GDGGG G G G GGG
Sbjct: 348 GGGGEGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGGGDL**YGGGGDGDGGG 175
Score = 41.2 bits (95), Expect = 7e-05
Identities = 26/60 (43%), Positives = 29/60 (48%), Gaps = 7/60 (11%)
Frame = -1
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS---GGGGGGGDGGGN 61
GG DG G D G GGD GGGG +G+ GGD GGGG G GGG+
Sbjct: 156 GGGGDGDGGGGDL***GGGGDGSGGGG---------DGEGGGGDL**YGGGGEGDGGGGD 4
Score = 35.4 bits (80), Expect = 0.004
Identities = 27/58 (46%), Positives = 28/58 (47%), Gaps = 8/58 (13%)
Frame = -1
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGG------NGCGSGGGGG 70
G GGD GGGG + GD GG GGGG GDGGG G G G GGG
Sbjct: 444 GGGGDDVGGGGDL***GG*ETGDGGGGLL***GGGGEGDGGGGDL***GGGGDGDGGG 271
Score = 22.7 bits (47), Expect(2) = 2e-06
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = -3
Query: 76 WWRRRWWR 83
WW RWWR
Sbjct: 151 WW*WRWWR 128
>TC208205 hydroxyproline-rich glycoprotein
Length = 603
Score = 52.8 bits (125), Expect = 2e-08
Identities = 33/70 (47%), Positives = 38/70 (54%), Gaps = 6/70 (8%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD--SGGGGGGGDGGGNG 62
GG +G G D G GG+ DGGGG + EGD GGD + GGGG GDGGG
Sbjct: 305 GGGGEGDGGGGDL**YGEGGEGDGGGGDLYT*GGGGEGDGGGGDLYT*GGGGDGDGGGGD 126
Query: 63 CGSGGGGGCG 72
+ GGGG G
Sbjct: 125 L*TYGGGGEG 96
Score = 52.8 bits (125), Expect = 2e-08
Identities = 33/68 (48%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS--GGGGGGGDGGGNG 62
GG +G G D G GGD DGGGG + EGD GGD GGGG GDGGG
Sbjct: 209 GGGGEGDGGGGDLYT*GGGGDGDGGGGDL*TYGGGGEGDGGGGDL*IYGGGGDGDGGGGD 30
Query: 63 CGSGGGGG 70
+ GGGG
Sbjct: 29 L*TYGGGG 6
Score = 51.2 bits (121), Expect = 7e-08
Identities = 34/71 (47%), Positives = 39/71 (54%), Gaps = 6/71 (8%)
Frame = -2
Query: 8 DGGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD--SGGGGGGGDGGGN 61
+GG DGG G D G GG+ DGGGG + +GD GGD + GGGG GDGGG
Sbjct: 254 EGGEGDGG--GGDLYT*GGGGEGDGGGGDLYT*GGGGDGDGGGGDL*TYGGGGEGDGGGG 81
Query: 62 GCGSGGGGGCG 72
GGGG G
Sbjct: 80 DL*IYGGGGDG 48
Score = 47.4 bits (111), Expect = 1e-06
Identities = 35/74 (47%), Positives = 37/74 (49%), Gaps = 13/74 (17%)
Frame = -2
Query: 9 GGFADGGSSGD---DGCGGDSDGGGGVVIEKVVMVEGDCSGGD----SGGGGGGGDG--- 58
GG DGG GD G GG+ DGGGG + EGD GGD GGG G G G
Sbjct: 347 GGECDGGG-GDL**YGGGGEGDGGGGDL**YGEGGEGDGGGGDLYT*GGGGEGDGGGGDL 171
Query: 59 ---GGNGCGSGGGG 69
GG G G GGGG
Sbjct: 170 YT*GGGGDGDGGGG 129
Score = 42.4 bits (98), Expect = 3e-05
Identities = 28/59 (47%), Positives = 30/59 (50%), Gaps = 10/59 (16%)
Frame = -2
Query: 21 GCGGDSDGGGGVVIEKVVMVEGDCSGGD-----SGGGGGGGDG-----GGNGCGSGGGG 69
G GG+ DGGGG + EGD GGD GG G GG G GG G G GGGG
Sbjct: 353 GGGGECDGGGGDL**YGGGGEGDGGGGDL**YGEGGEGDGGGGDLYT*GGGGEGDGGGG 177
Score = 38.5 bits (88), Expect = 5e-04
Identities = 24/53 (45%), Positives = 27/53 (50%), Gaps = 4/53 (7%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGD 57
GG DG G D G GG+ DGGGG + +GD GGD GGGGD
Sbjct: 161 GGGGDGDGGGGDL*TYGGGGEGDGGGGDL*IYGGGGDGDGGGGDL*TYGGGGD 3
Score = 28.1 bits (61), Expect = 0.62
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 10/45 (22%)
Frame = -2
Query: 36 KVVMVEGDCSGGDSG----GGGGGGDGGG------NGCGSGGGGG 70
K++ ++G +GG GGGG DGGG G G G GGG
Sbjct: 410 KLI*LQG*YTGGGGEL*R*GGGGECDGGGGDL**YGGGGEGDGGG 276
>TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wall
structural protein 1.0 precursor (GRP 1.0), partial
(27%)
Length = 447
Score = 52.4 bits (124), Expect = 3e-08
Identities = 28/64 (43%), Positives = 30/64 (46%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
GG GG G +G G + GG G G GG GGGGG G GG G G G G
Sbjct: 18 GGAHGGGYGGGEGAGQGAGGGYG----------GGAVGGGGGGGGGSGGGGARGGGYGAG 167
Query: 69 GGCG 72
GG G
Sbjct: 168GGSG 179
Score = 41.2 bits (95), Expect = 7e-05
Identities = 17/27 (62%), Positives = 18/27 (65%)
Frame = +3
Query: 47 GDSGGGGGGGDGGGNGCGSGGGGGCGG 73
G +GG GGG GGG G G G GGG GG
Sbjct: 9 GGAGGAHGGGYGGGEGAGQGAGGGYGG 89
Score = 40.4 bits (93), Expect = 1e-04
Identities = 17/29 (58%), Positives = 19/29 (64%)
Frame = +3
Query: 45 SGGDSGGGGGGGDGGGNGCGSGGGGGCGG 73
+GG GGG GGG+G G G G G GGG G
Sbjct: 15 AGGAHGGGYGGGEGAGQGAGGGYGGGAVG 101
Score = 35.8 bits (81), Expect = 0.003
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 5/37 (13%)
Frame = +3
Query: 42 GDCSGGDSGGGGGG-----GDGGGNGCGSGGGGGCGG 73
G G GG GGG G GGG G G+ GGGG GG
Sbjct: 9 GGAGGAHGGGYGGGEGAGQGAGGGYGGGAVGGGGGGG 119
Score = 32.7 bits (73), Expect = 0.025
Identities = 22/59 (37%), Positives = 24/59 (40%)
Frame = +3
Query: 6 VDDGGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCG 64
V GG GGS GGGG GG G GGG G+GGG+G G
Sbjct: 96 VGGGGGGGGGS-----------GGGG------------ARGGGYGAGGGSGEGGGHGGG 203
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 52.4 bits (124), Expect = 3e-08
Identities = 29/65 (44%), Positives = 30/65 (45%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGG 68
G + GG G G GG GG GG GG GGGG GGG G G GGG
Sbjct: 78 GSYGXGGGRGGYGGGGRGGRGG--------------YGGGGGGYGGGGYGGGGGYGGGGG 215
Query: 69 GGCGG 73
GG GG
Sbjct: 216 GGGGG 230
Score = 51.2 bits (121), Expect = 7e-08
Identities = 28/65 (43%), Positives = 32/65 (49%), Gaps = 6/65 (9%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIE--KVVMVEGDCSGGDSGGGGGGGDGGGNG---- 62
GG+ GG G G GG GGGG + + + DCS G GGG GG GGG G
Sbjct: 159 GGYGGGGYGGGGGYGGGGGGGGGGCYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYN 338
Query: 63 CGSGG 67
CG G
Sbjct: 339 CGESG 353
Score = 48.9 bits (115), Expect = 3e-07
Identities = 32/80 (40%), Positives = 33/80 (41%), Gaps = 15/80 (18%)
Frame = +3
Query: 9 GGFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGC----- 63
GG+ GG G G GG G GG G GG GG GGGG GGG GC
Sbjct: 105 GGYGGGGRGGRGGYGGGGGGYGG----------GGYGGG--GGYGGGGGGGGGGCYKCGE 248
Query: 64 ----------GSGGGGGCGG 73
G GGGG GG
Sbjct: 249 TGHIARDCSQGGGGGGRYGG 308
Score = 35.8 bits (81), Expect = 0.003
Identities = 25/57 (43%), Positives = 25/57 (43%), Gaps = 1/57 (1%)
Frame = +3
Query: 18 GDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDG-GGNGCGSGGGGGCGG 73
G G GG G G G GG GGG GG G GG G G GGGG GG
Sbjct: 48 GTRGGGGXXRGSYGX---------GGGRGGYGGGGRGGRGGYGGGGGGYGGGGYGGG 191
Score = 25.8 bits (55), Expect = 3.1
Identities = 15/34 (44%), Positives = 15/34 (44%), Gaps = 1/34 (2%)
Frame = +3
Query: 41 EGDCSGGDSGGGGGGGDGGGNGCGSG-GGGGCGG 73
E G GGG G G G G GGGG GG
Sbjct: 33 ESPVQGTRGGGGXXRGSYGXGGGRGGYGGGGRGG 134
>BU763328 similar to SP|P10496|GRP2_ Glycine-rich cell wall structural
protein 1.8 precursor (GRP 1.8). [Kidney bean French
bean], partial (11%)
Length = 169
Score = 52.4 bits (124), Expect = 3e-08
Identities = 28/64 (43%), Positives = 33/64 (50%)
Frame = -3
Query: 10 GFADGGSSGDDGCGGDSDGGGGVVIEKVVMVEGDCSGGDSGGGGGGGDGGGNGCGSGGGG 69
G+ DGG+ G GG G GG G +GG+ GGG GGG+GGG G G G G
Sbjct: 167 GYGDGGAHGGGYGGGQGGGAGG----------GYGAGGEHGGGYGGGEGGGAGGGYGASG 18
Query: 70 GCGG 73
GG
Sbjct: 17 EHGG 6
>TC224645 homologue to UP|Q41707 (Q41707) Extensin class 1 protein precursor,
partial (58%)
Length = 856
Score = 52.4 bits (124), Expect = 3e-08
Identities = 36/75 (48%), Positives = 38/75 (50%), Gaps = 14/75 (18%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDG---- 58
GG DG G D G GGD DGGGG + +GD GGD GGGG GDG
Sbjct: 753 GGGGDGDGGGGDL***GGGGDGDGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGD 574
Query: 59 ----GGNGCGSGGGG 69
GG G GSGGGG
Sbjct: 573 L***GGGGDGSGGGG 529
Score = 51.2 bits (121), Expect = 7e-08
Identities = 35/74 (47%), Positives = 37/74 (49%), Gaps = 12/74 (16%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGG-- 60
GG DG G D G GGD DGGGG + +GD GGD GGGG GDGGG
Sbjct: 465 GGGGDGDGGGGDL***GGGGDGDGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGD 286
Query: 61 ----NGCGSGGGGG 70
G G G GGG
Sbjct: 285 L***GGGGDGDGGG 244
Score = 50.8 bits (120), Expect = 9e-08
Identities = 35/74 (47%), Positives = 37/74 (49%), Gaps = 12/74 (16%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGG-- 60
GG DG G D G GGD DGGGG + +GD GGD GGGG GDGGG
Sbjct: 561 GGGGDGSGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGGGD 382
Query: 61 ----NGCGSGGGGG 70
G G G GGG
Sbjct: 381 L**YGGGGDGDGGG 340
Score = 50.8 bits (120), Expect = 9e-08
Identities = 38/88 (43%), Positives = 41/88 (46%), Gaps = 15/88 (17%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGGN- 61
GG DG G D G GGD DGGGG + +GD GGD GGGG GDGGG
Sbjct: 417 GGGGDGDGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGGGD 238
Query: 62 ----GCGSGGGGGCGGL----MWWRRRW 81
G G G GG G L W+ W
Sbjct: 237 L***GGGGDGDGGGGDL***GGWYGSLW 154
Score = 50.8 bits (120), Expect = 9e-08
Identities = 35/74 (47%), Positives = 37/74 (49%), Gaps = 12/74 (16%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS--GGGGGGGDGGG-- 60
GG DG G D G GGD DGGGG + +GD GGD GGGG GDGGG
Sbjct: 513 GGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGGGDL**YGGGGDGDGGGGD 334
Query: 61 ----NGCGSGGGGG 70
G G G GGG
Sbjct: 333 L***GGGGDGDGGG 292
Score = 49.7 bits (117), Expect = 2e-07
Identities = 34/75 (45%), Positives = 36/75 (47%), Gaps = 14/75 (18%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGD----SGGGGGGGDG-- 58
GG DG G D G GGD DGGGG + +GD GGD GGG G G G
Sbjct: 705 GGGGDGDGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGSGGGGD 526
Query: 59 ----GGNGCGSGGGG 69
GG G G GGGG
Sbjct: 525 L**YGGGGDGDGGGG 481
Score = 49.3 bits (116), Expect = 3e-07
Identities = 34/74 (45%), Positives = 37/74 (49%), Gaps = 12/74 (16%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS--GGGGGGGDGGG-- 60
GG +G G D G GGD DGGGG + +GD GGD GGGG GDGGG
Sbjct: 801 GGGGEGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGGGDL**YGGGGDGDGGGGD 622
Query: 61 ----NGCGSGGGGG 70
G G G GGG
Sbjct: 621 L***GGGGDGDGGG 580
Score = 48.9 bits (115), Expect = 3e-07
Identities = 34/74 (45%), Positives = 36/74 (47%), Gaps = 12/74 (16%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGG-- 60
GG DG G D G GGD GGGG + +GD GGD GGGG GDGGG
Sbjct: 609 GGGGDGDGGGGDL***GGGGDGSGGGGDL**YGGGGDGDGGGGDL***GGGGDGDGGGGD 430
Query: 61 ----NGCGSGGGGG 70
G G G GGG
Sbjct: 429 L***GGGGDGDGGG 388
Score = 48.5 bits (114), Expect = 4e-07
Identities = 34/74 (45%), Positives = 36/74 (47%), Gaps = 12/74 (16%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS--GGGGGGGDGGG-- 60
GG DG G D G GGD DGGGG + +G GGD GGGG GDGGG
Sbjct: 657 GGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGSGGGGDL**YGGGGDGDGGGGD 478
Query: 61 ----NGCGSGGGGG 70
G G G GGG
Sbjct: 477 L***GGGGDGDGGG 436
Score = 40.4 bits (93), Expect = 1e-04
Identities = 33/89 (37%), Positives = 36/89 (40%), Gaps = 24/89 (26%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVVIEKVVMVEGDCSGGDS--------------- 49
GG DG G D G GGD DGGGG + +GD GGD
Sbjct: 321 GGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGGGDL***GGWYGSLWGGGG 142
Query: 50 -----GGGGGGGDGGGNGCGSGGGGGCGG 73
G GG G GGG+ GGG G GG
Sbjct: 141 DL***GEGGDGDGGGGDL**YGGGDGDGG 55
Score = 38.9 bits (89), Expect(2) = 3e-05
Identities = 26/52 (50%), Positives = 28/52 (53%), Gaps = 8/52 (15%)
Frame = -2
Query: 27 DGGGGVVIEKVVMVEGDCSGGDSG--GGGGGGDGGG------NGCGSGGGGG 70
DGGGG++ EGD GGD GGGG GDGGG G G G GGG
Sbjct: 831 DGGGGLL***GGGGEGDGGGGDL***GGGGDGDGGGGDL***GGGGDGDGGG 676
Score = 36.6 bits (83), Expect = 0.002
Identities = 34/85 (40%), Positives = 36/85 (42%), Gaps = 24/85 (28%)
Frame = -2
Query: 9 GGFADGGSSGDD----GCGGDSDGGGGVV------IEKVVMVEGDC----SGGDSGGGGG 54
GG DG G D G GGD DGGGG + + GD GGD GGGG
Sbjct: 273 GGGGDGDGGGGDL***GGGGDGDGGGGDL***GGWYGSLWGGGGDL***GEGGDGDGGGG 94
Query: 55 ------GGD-GGGNGC---GSGGGG 69
GGD GG G GGGG
Sbjct: 93 DL**YGGGDGDGGGGLL***EGGGG 19
Score = 26.6 bits (57), Expect = 1.8
Identities = 15/29 (51%), Positives = 18/29 (61%), Gaps = 3/29 (10%)
Frame = -2
Query: 8 DGGFADGGSSGDD---GCGGDSDGGGGVV 33
+GG DGG G D GGD DGGGG++
Sbjct: 123 EGGDGDGG--GGDL**YGGGDGDGGGGLL 43
Score = 22.7 bits (47), Expect(2) = 3e-05
Identities = 6/8 (75%), Positives = 6/8 (75%)
Frame = -1
Query: 76 WWRRRWWR 83
WW RWWR
Sbjct: 604 WW*WRWWR 581
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.145 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,908,001
Number of Sequences: 63676
Number of extensions: 149073
Number of successful extensions: 15552
Number of sequences better than 10.0: 1787
Number of HSP's better than 10.0 without gapping: 4324
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8667
length of query: 117
length of database: 12,639,632
effective HSP length: 93
effective length of query: 24
effective length of database: 6,717,764
effective search space: 161226336
effective search space used: 161226336
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0072a.11


