

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0069.6
(167 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
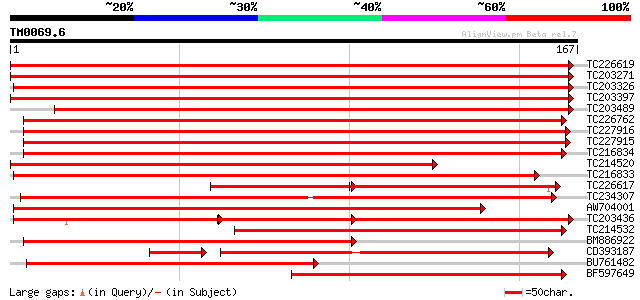
Score E
Sequences producing significant alignments: (bits) Value
TC226619 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxid... 305 9e-84
TC203271 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid... 261 1e-70
TC203326 similar to UP|GPX4_ARATH (O48646) Probable phospholipid... 258 7e-70
TC203397 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid... 258 1e-69
TC203489 similar to UP|GPX4_ARATH (O48646) Probable phospholipid... 239 6e-64
TC226762 similar to UP|GPX1_PEA (O24296) Phospholipid hydroperox... 232 5e-62
TC227916 weakly similar to UP|GPX3_ARATH (Q9LYB4) Probable gluta... 217 2e-57
TC227915 weakly similar to UP|GPX3_ARATH (Q9LYB4) Probable gluta... 216 5e-57
TC216834 similar to GB|AAO39963.1|28372962|BT003735 At2g48150 {A... 208 8e-55
TC214520 similar to UP|Q8W2G9 (Q8W2G9) Glutathione peroxidase, p... 185 1e-47
TC216833 similar to GB|AAO39963.1|28372962|BT003735 At2g48150 {A... 179 7e-46
TC226617 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxid... 112 2e-44
TC234307 weakly similar to UP|GPX4_MESCR (Q9LEF0) Probable phosp... 170 3e-43
AW704001 weakly similar to GP|21360380|gb glutathione peroxidase... 165 8e-42
TC203436 similar to UP|Q6QHC9 (Q6QHC9) Phospholipid-hydroperoxid... 104 1e-39
TC214532 similar to GB|AAO23624.1|27765006|BT003059 At1g63460 {A... 145 9e-36
BM886922 weakly similar to GP|28372962|gb At2g48150 {Arabidopsis... 131 2e-31
CD393187 108 4e-27
BU761482 similar to GP|28372962|gb| At2g48150 {Arabidopsis thali... 112 8e-26
BF597649 similar to PIR|S33618|S336 glutathione peroxidase (EC 1... 110 3e-25
>TC226619 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxide
glutathione peroxidase, complete
Length = 956
Score = 305 bits (780), Expect = 9e-84
Identities = 143/166 (86%), Positives = 160/166 (96%)
Frame = +3
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MAEQ+S S+YDFTVKDI GNDVSL+ YSGKVL+IVNVASQCGLTQTNYKELN+LYEKYK+
Sbjct: 219 MAEQSSNSIYDFTVKDISGNDVSLNDYSGKVLLIVNVASQCGLTQTNYKELNVLYEKYKN 398
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
+G EILAFPCNQFAGQEPG N+EIQ+VVCTRFK+EFP+FDKVEVNGKNA PL+KFLK++K
Sbjct: 399 QGFEILAFPCNQFAGQEPGNNEEIQEVVCTRFKAEFPIFDKVEVNGKNAVPLYKFLKEKK 578
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQS 166
GGIFGDGIKWNFTKFLVNKEGKVV+RYAPTTSP+KIEKD+EKLLQS
Sbjct: 579 GGIFGDGIKWNFTKFLVNKEGKVVDRYAPTTSPLKIEKDIEKLLQS 716
>TC203271 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid
hydroperoxide glutathione peroxidase (PHGPx)
(Salt-associated protein) , partial (98%)
Length = 778
Score = 261 bits (666), Expect = 1e-70
Identities = 121/166 (72%), Positives = 144/166 (85%)
Frame = +2
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MA ++KS++DFTVKD +GND++L Y GKVLIIVNVASQCGLT +NY EL+ LYEKYK
Sbjct: 95 MATSSAKSVHDFTVKDAKGNDINLGDYKGKVLIIVNVASQCGLTNSNYTELSQLYEKYKQ 274
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
KGLEILAFPCNQF QEPG+N++IQ+ VCTRFK+EFPVFDKV+VNG A PL+K+LK K
Sbjct: 275 KGLEILAFPCNQFGAQEPGSNEQIQEFVCTRFKAEFPVFDKVDVNGDKAAPLYKYLKSSK 454
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQS 166
GG+FGDGIKWNF+KFLV+KEG VV+RYAPTTSP+ IEKDL KLL +
Sbjct: 455 GGLFGDGIKWNFSKFLVDKEGNVVDRYAPTTSPLSIEKDLLKLLDA 592
>TC203326 similar to UP|GPX4_ARATH (O48646) Probable phospholipid
hydroperoxide glutathione peroxidase, mitochondrial
precursor (PHGPx) (AtGPX1) , partial (74%)
Length = 749
Score = 258 bits (660), Expect = 7e-70
Identities = 118/165 (71%), Positives = 143/165 (86%)
Frame = +2
Query: 2 AEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSK 61
++ +KS++DFTVKD RGN+V+L+ Y GKVL+IVNVASQCGLT +NY ELN LYEKYK K
Sbjct: 56 SQSNTKSVHDFTVKDARGNNVNLADYKGKVLLIVNVASQCGLTNSNYTELNQLYEKYKGK 235
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKG 121
GLEILAFPCNQF QEPGTN+EI + CTRFK+EFP+FDKV+VNG NA PL+KFLK KG
Sbjct: 236 GLEILAFPCNQFGAQEPGTNEEIVEFACTRFKAEFPIFDKVDVNGDNAAPLYKFLKSSKG 415
Query: 122 GIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQS 166
G+FGD IKWNF+KFLV+K+G VV+RYAPTTSP+ IEKD++KLL +
Sbjct: 416 GLFGDSIKWNFSKFLVDKDGNVVDRYAPTTSPLSIEKDIKKLLDA 550
>TC203397 similar to UP|GPX4_CITSI (Q06652) Probable phospholipid
hydroperoxide glutathione peroxidase (PHGPx)
(Salt-associated protein) , partial (94%)
Length = 1010
Score = 258 bits (658), Expect = 1e-69
Identities = 120/166 (72%), Positives = 141/166 (84%)
Frame = +3
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
MA +KS++DFTVKD +GND++L Y GKVLIIVNVASQCGLT +NY EL+ LYEKYK
Sbjct: 327 MATSNAKSVHDFTVKDAKGNDINLGDYKGKVLIIVNVASQCGLTNSNYTELSQLYEKYKQ 506
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
KGLEILAFPCNQF QEPG+N++IQ+ VCTRFK+EFPVFDKV+VNG A PL+K+LK K
Sbjct: 507 KGLEILAFPCNQFGAQEPGSNEQIQEFVCTRFKAEFPVFDKVDVNGDKAAPLYKYLKSSK 686
Query: 121 GGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQS 166
GG+ GDGIKWNF KFLV+KEG VV+RYAPTTSP+ IEKDL KLL +
Sbjct: 687 GGLLGDGIKWNFAKFLVDKEGNVVDRYAPTTSPLSIEKDLLKLLDA 824
>TC203489 similar to UP|GPX4_ARATH (O48646) Probable phospholipid
hydroperoxide glutathione peroxidase, mitochondrial
precursor (PHGPx) (AtGPX1) , partial (65%)
Length = 606
Score = 239 bits (609), Expect = 6e-64
Identities = 110/153 (71%), Positives = 130/153 (84%)
Frame = +1
Query: 14 VKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEILAFPCNQF 73
VKD RGNDV+L+ VL++VNVASQCGLT +NY ELN LYEKYK KGLEILAFPCNQF
Sbjct: 4 VKDARGNDVNLADXXXXVLLLVNVASQCGLTNSNYTELNQLYEKYKGKGLEILAFPCNQF 183
Query: 74 AGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFT 133
QEPGTN+EI + CTRFK+EFP+FDKV+VNG NA PL+KFLK KGG+FGD IKWNF+
Sbjct: 184 GAQEPGTNEEIVEFACTRFKAEFPIFDKVDVNGDNAAPLYKFLKSSKGGLFGDSIKWNFS 363
Query: 134 KFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQS 166
KFLV+K+G VV+RYAPTTSP+ IEKD++KLL +
Sbjct: 364 KFLVDKDGNVVDRYAPTTSPLSIEKDIKKLLDA 462
>TC226762 similar to UP|GPX1_PEA (O24296) Phospholipid hydroperoxide
glutathione peroxidase, chloroplast precursor (PHGPx) ,
partial (90%)
Length = 1008
Score = 232 bits (592), Expect = 5e-62
Identities = 107/160 (66%), Positives = 133/160 (82%)
Frame = +1
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
T K++YDFTVKDI DVSLS++ GKVL+IVNVAS+CGLT +NY EL+ LYEKYK++GLE
Sbjct: 244 TEKTIYDFTVKDIDRKDVSLSKFKGKVLLIVNVASRCGLTSSNYSELSRLYEKYKNQGLE 423
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIF 124
ILAFPCNQF QEPG+N++I+ CTR+K+EFP+FDKV+VNG P+++FLK GG
Sbjct: 424 ILAFPCNQFGMQEPGSNEDIKQFACTRYKAEFPIFDKVDVNGPFTTPVYQFLKSSAGGFL 603
Query: 125 GDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
GD IKWNF KFLV+K GKV+ERY PTTSP +IEKD++KLL
Sbjct: 604 GDLIKWNFEKFLVDKNGKVIERYPPTTSPFQIEKDIQKLL 723
>TC227916 weakly similar to UP|GPX3_ARATH (Q9LYB4) Probable glutathione
peroxidase At3g63080 , partial (95%)
Length = 1013
Score = 217 bits (553), Expect = 2e-57
Identities = 95/161 (59%), Positives = 126/161 (78%)
Frame = +1
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
+ KS ++FTVKD RG DV+L+ Y GKVL+++NVAS+CG NY +L LY YKS+GLE
Sbjct: 193 SEKSFHEFTVKDARGKDVNLNAYRGKVLLVINVASKCGFADANYTQLTQLYSTYKSRGLE 372
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIF 124
ILAFPCNQF +EPGT+ E QD CTR+K+E+P+F K+ VNG + P+FKFLK QK G+
Sbjct: 373 ILAFPCNQFLKKEPGTSQEAQDFACTRYKAEYPIFGKIRVNGSDTAPVFKFLKTQKSGVM 552
Query: 125 GDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQ 165
G IKWNFTKFLV++EG+V++RY+PTT P+ IE D++K L+
Sbjct: 553 GSRIKWNFTKFLVDEEGRVIQRYSPTTKPLAIENDIKKALR 675
>TC227915 weakly similar to UP|GPX3_ARATH (Q9LYB4) Probable glutathione
peroxidase At3g63080 , partial (95%)
Length = 885
Score = 216 bits (549), Expect = 5e-57
Identities = 93/161 (57%), Positives = 126/161 (77%)
Frame = +1
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
+ S+++FTVKD RG DV+L+ Y GKVL+++NVAS+CG NY +L +Y YKS+GLE
Sbjct: 124 SENSIHEFTVKDARGKDVNLNAYRGKVLLVINVASKCGFADANYSQLTQIYSTYKSRGLE 303
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIF 124
ILAFPCNQF +EPGT+ E Q+ CTR+K+E+P+F K+ VNG + P+FKFLK QK G+
Sbjct: 304 ILAFPCNQFLKKEPGTSQEAQEFACTRYKAEYPIFGKIRVNGSDTAPVFKFLKTQKSGVM 483
Query: 125 GDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLLQ 165
G IKWNFTKFLV++EG+V++RY+PTT P+ IE D++K LQ
Sbjct: 484 GSRIKWNFTKFLVDEEGRVIQRYSPTTKPLAIESDIKKALQ 606
>TC216834 similar to GB|AAO39963.1|28372962|BT003735 At2g48150 {Arabidopsis
thaliana;} , partial (98%)
Length = 821
Score = 208 bits (530), Expect = 8e-55
Identities = 93/160 (58%), Positives = 122/160 (76%)
Frame = +3
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
+ KS+++F VKD +G DV+LS Y GKVL++VNVAS+CG T TNY +L LY KYK +GLE
Sbjct: 132 SEKSIHEFMVKDAKGRDVNLSIYKGKVLLVVNVASKCGFTNTNYTQLTELYSKYKDRGLE 311
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIF 124
ILAFPCNQF QEPG++ ++++ CTR+K+ +P+F KV VNG + P++KFLK K G
Sbjct: 312 ILAFPCNQFLKQEPGSSQDVEEFACTRYKAAYPIFGKVRVNGPDTAPVYKFLKANKSGFL 491
Query: 125 GDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
G IKWNFTKFLV+KEG V+ RY PTTSP+ IE D++ L
Sbjct: 492 GTRIKWNFTKFLVDKEGNVLPRYGPTTSPLSIENDIKTAL 611
>TC214520 similar to UP|Q8W2G9 (Q8W2G9) Glutathione peroxidase, partial (69%)
Length = 415
Score = 185 bits (469), Expect = 1e-47
Identities = 88/126 (69%), Positives = 103/126 (80%)
Frame = +2
Query: 1 MAEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKS 60
M + KS+YDF VKD +G+DV LS Y GKVL+IVNVASQCGLT +NY ELN LY+KYK
Sbjct: 38 MTTKDPKSVYDFVVKDAKGDDVDLSFYKGKVLLIVNVASQCGLTNSNYTELNQLYDKYKD 217
Query: 61 KGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQK 120
+GLEILAFPCNQF QEP +ND+I D VC+RFKSEFP+FDK+EVNG N+ PL+KFLK K
Sbjct: 218 QGLEILAFPCNQFGKQEPESNDKIVDFVCSRFKSEFPIFDKIEVNGDNSAPLYKFLKLGK 397
Query: 121 GGIFGD 126
GIFGD
Sbjct: 398 WGIFGD 415
>TC216833 similar to GB|AAO39963.1|28372962|BT003735 At2g48150 {Arabidopsis
thaliana;} , partial (80%)
Length = 538
Score = 179 bits (453), Expect = 7e-46
Identities = 84/155 (54%), Positives = 108/155 (69%)
Frame = +2
Query: 2 AEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSK 61
A T KS+++F VKD +G DV+LS Y GKVL++VNVAS+CG T +NY +L LY KYK +
Sbjct: 65 ASVTEKSIHEFMVKDAKGRDVNLSTYKGKVLLVVNVASKCGFTNSNYTQLTELYSKYKDR 244
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKG 121
GLEILAFPCNQF QEPG++ E ++ CTR+K+E+P+F KV VNG + P++KFLK K
Sbjct: 245 GLEILAFPCNQFLKQEPGSSQEAEEFACTRYKAEYPIFGKVRVNGPDTAPVYKFLKANKT 424
Query: 122 GIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKI 156
G G IK N T V+ EG V+ Y TT P I
Sbjct: 425 GFLGSRIKGNLTGLWVDYEGHVLASYGSTTLPFAI 529
>TC226617 similar to UP|Q8W259 (Q8W259) Phospholipid hydroperoxide
glutathione peroxidase, partial (59%)
Length = 871
Score = 112 bits (281), Expect(2) = 2e-44
Identities = 53/63 (84%), Positives = 60/63 (95%), Gaps = 1/63 (1%)
Frame = +2
Query: 101 KVEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIE-KD 159
+VEVNGKNA PL+KFLK+QKGGIFGDGIKWNFTKFLVNKEGKVV+RYAPTTSP+KIE KD
Sbjct: 227 QVEVNGKNAAPLYKFLKEQKGGIFGDGIKWNFTKFLVNKEGKVVDRYAPTTSPLKIEVKD 406
Query: 160 LEK 162
+ +
Sbjct: 407 IRE 415
Score = 82.8 bits (203), Expect(2) = 2e-44
Identities = 36/43 (83%), Positives = 40/43 (92%)
Frame = +3
Query: 60 SKGLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKV 102
S G EILAFPCNQFAGQEPG N+EIQ+VVCTRFK+EFP+FDKV
Sbjct: 72 SPGFEILAFPCNQFAGQEPGNNEEIQEVVCTRFKAEFPIFDKV 200
>TC234307 weakly similar to UP|GPX4_MESCR (Q9LEF0) Probable phospholipid
hydroperoxide glutathione peroxidase (PHGPx) , partial
(66%)
Length = 558
Score = 170 bits (431), Expect = 3e-43
Identities = 78/158 (49%), Positives = 115/158 (72%)
Frame = +2
Query: 4 QTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGL 63
+ ++++Y+F D+ G VS+ +Y KVLI+VNVA+ CGLT+ NY +LN +Y +YK +GL
Sbjct: 35 KNAETVYEFEATDLDGAPVSMDKYKDKVLIVVNVATYCGLTRANYAQLNDIYNEYKDRGL 214
Query: 64 EILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGI 123
EI FPCNQF QEPG + +I++ + + K + V+DKV+VNGK+A PL+K+L+ +KGG
Sbjct: 215 EIAGFPCNQFGNQEPGCSVDIKEFL-LKNKVTWDVYDKVDVNGKDAHPLWKWLQHKKGGF 391
Query: 124 FGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLE 161
+GIKWNFTKFL++++G V+RYAPT P D+E
Sbjct: 392 ITNGIKWNFTKFLIDRKGNPVKRYAPTDEPKTFVTDIE 505
>AW704001 weakly similar to GP|21360380|gb glutathione peroxidase 1 {Oryza
sativa}, partial (80%)
Length = 424
Score = 165 bits (418), Expect = 8e-42
Identities = 82/139 (58%), Positives = 102/139 (72%)
Frame = +2
Query: 2 AEQTSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSK 61
A ++K+ +D TVKD +G D++L GK LI+V VASQCGLT +NY EL+ EKYK K
Sbjct: 2 ATSSAKADHDDTVKDAKGKDINLGDCEGKELIMVKVASQCGLTNSNYTELSQRCEKYKQK 181
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKG 121
GLE LAFP NQ QEPG+N+++Q VCTRFK+E P DKV+VNG A PL K+LK G
Sbjct: 182 GLEKLAFPYNQSGAQEPGSNEQMQ*SVCTRFKAE*PELDKVDVNGDEAAPLCKYLKVNTG 361
Query: 122 GIFGDGIKWNFTKFLVNKE 140
G+FGDGIKWNF KF V++E
Sbjct: 362 GLFGDGIKWNFCKFRVDEE 418
>TC203436 similar to UP|Q6QHC9 (Q6QHC9) Phospholipid-hydroperoxide
glutathione peroxidase , partial (97%)
Length = 1109
Score = 104 bits (260), Expect(2) = 1e-39
Identities = 46/65 (70%), Positives = 57/65 (86%)
Frame = +1
Query: 102 VEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLE 161
V+VNG NA PL+KFLK KGG+FGD IKWNF+KFLV+K+G VV+RYAPTTSP+ IEKD++
Sbjct: 694 VDVNGDNAAPLYKFLKSSKGGLFGDSIKWNFSKFLVDKDGNVVDRYAPTTSPLSIEKDIK 873
Query: 162 KLLQS 166
KLL +
Sbjct: 874 KLLDA 888
Score = 80.5 bits (197), Expect = 3e-16
Identities = 42/86 (48%), Positives = 53/86 (60%), Gaps = 24/86 (27%)
Frame = +3
Query: 2 AEQTSKSLYDFTVK------------------------DIRGNDVSLSQYSGKVLIIVNV 37
++ +KS++DFTVK D RGNDV+L+ Y GKVL++VNV
Sbjct: 60 SQSNTKSVHDFTVKVCLLTH**FPLVYLLDQNDYVCLQDARGNDVNLADYKGKVLLLVNV 239
Query: 38 ASQCGLTQTNYKELNILYEKYKSKGL 63
ASQCGLT +NY ELN LYEKYK KG+
Sbjct: 240 ASQCGLTNSNYTELNQLYEKYKGKGM 317
Score = 75.1 bits (183), Expect(2) = 1e-39
Identities = 32/41 (78%), Positives = 36/41 (87%)
Frame = +2
Query: 62 GLEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKV 102
GLEILAFPCNQF QEPGTN+EI + CTRFK+EFP+FDKV
Sbjct: 497 GLEILAFPCNQFGAQEPGTNEEIVEFACTRFKAEFPIFDKV 619
>TC214532 similar to GB|AAO23624.1|27765006|BT003059 At1g63460 {Arabidopsis
thaliana;} , partial (60%)
Length = 825
Score = 145 bits (366), Expect = 9e-36
Identities = 65/98 (66%), Positives = 82/98 (83%)
Frame = +1
Query: 67 AFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGD 126
AFPCNQF QEP +ND+I D VC+RFKSEFP+FDK+EVNG N+ PL+KFLK K GIFGD
Sbjct: 370 AFPCNQFGKQEPESNDKIVDFVCSRFKSEFPIFDKIEVNGDNSAPLYKFLKLGKWGIFGD 549
Query: 127 GIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDLEKLL 164
I+WNF+KF+V+K G+VV RY PTTSP+ +E+D+ +LL
Sbjct: 550 DIQWNFSKFVVDKNGQVVGRYYPTTSPLSLERDIHQLL 663
>BM886922 weakly similar to GP|28372962|gb At2g48150 {Arabidopsis thaliana},
partial (61%)
Length = 423
Score = 131 bits (329), Expect = 2e-31
Identities = 56/98 (57%), Positives = 76/98 (77%)
Frame = +2
Query: 5 TSKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLE 64
+ S+++FTVKD RG DV+L+ Y GKVL+++NVAS+CG NY +L +Y YKS+GLE
Sbjct: 71 SENSIHEFTVKDARGKDVNLNAYRGKVLLVINVASKCGFADANYSQLTQIYSTYKSRGLE 250
Query: 65 ILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKV 102
ILAFPCNQF +EPGT+ E Q+ CTR+K+E+P+F KV
Sbjct: 251 ILAFPCNQFLKKEPGTSQEAQEFACTRYKAEYPIFGKV 364
>CD393187
Length = 566
Score = 108 bits (269), Expect(2) = 4e-27
Identities = 55/98 (56%), Positives = 68/98 (69%)
Frame = -2
Query: 63 LEILAFPCNQFAGQEPGTNDEIQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGG 122
LEILAFPCNQF QEP +ND+I D VC+ FKSEFP+F K G K G
Sbjct: 418 LEILAFPCNQFGKQEPESNDKIVDFVCSGFKSEFPIFHK--*MGTTLLLCTNS*SRGKWG 245
Query: 123 IFGDGIKWNFTKFLVNKEGKVVERYAPTTSPMKIEKDL 160
IFGD I+WNF+KF+V+K G+VV RY PTTSP+ +E+D+
Sbjct: 244 IFGDDIQWNFSKFVVDKNGQVVGRYYPTTSPLSLERDI 131
Score = 29.6 bits (65), Expect(2) = 4e-27
Identities = 12/17 (70%), Positives = 14/17 (81%)
Frame = -3
Query: 42 GLTQTNYKELNILYEKY 58
GLT +NY ELN LY+KY
Sbjct: 492 GLTNSNYTELNQLYDKY 442
>BU761482 similar to GP|28372962|gb| At2g48150 {Arabidopsis thaliana},
partial (54%)
Length = 445
Score = 112 bits (280), Expect = 8e-26
Identities = 49/86 (56%), Positives = 66/86 (75%)
Frame = +3
Query: 6 SKSLYDFTVKDIRGNDVSLSQYSGKVLIIVNVASQCGLTQTNYKELNILYEKYKSKGLEI 65
S++L+ + +D +G DV+LS Y GKVL++VNVAS+CG T +NY +L LY KYK +GLEI
Sbjct: 186 SRNLFSWLAQDAKGRDVNLSTYKGKVLLVVNVASKCGFTNSNYTQLTELYSKYKDRGLEI 365
Query: 66 LAFPCNQFAGQEPGTNDEIQDVVCTR 91
LAFPCNQF QEPG++ E ++ CTR
Sbjct: 366 LAFPCNQFLKQEPGSSQEAEEFACTR 443
>BF597649 similar to PIR|S33618|S336 glutathione peroxidase (EC 1.11.1.9) -
sweet orange, partial (48%)
Length = 247
Score = 110 bits (275), Expect = 3e-25
Identities = 53/81 (65%), Positives = 64/81 (78%)
Frame = -1
Query: 84 IQDVVCTRFKSEFPVFDKVEVNGKNAEPLFKFLKDQKGGIFGDGIKWNFTKFLVNKEGKV 143
I++ V TRFK +F VFDK +VNG A PL+K+LK KGG+FGDGIKWNF+KF V+KEG V
Sbjct: 247 IKEFV*TRFKVKFAVFDKGDVNGDKAVPLYKYLKSSKGGLFGDGIKWNFSKFFVDKEGNV 68
Query: 144 VERYAPTTSPMKIEKDLEKLL 164
V+ YA T SP+ IEKDL KLL
Sbjct: 67 VD*YATTNSPLSIEKDLLKLL 5
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,244,501
Number of Sequences: 63676
Number of extensions: 72000
Number of successful extensions: 293
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 284
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 287
length of query: 167
length of database: 12,639,632
effective HSP length: 90
effective length of query: 77
effective length of database: 6,908,792
effective search space: 531976984
effective search space used: 531976984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0069.6


