

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0069.1
(367 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
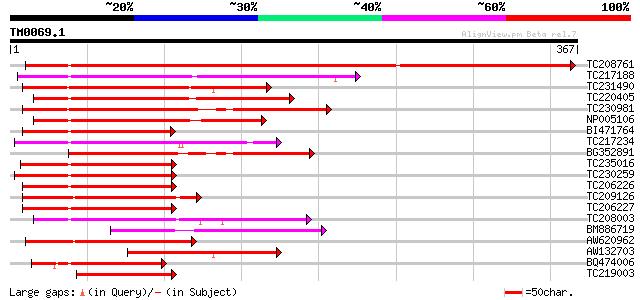
Score E
Sequences producing significant alignments: (bits) Value
TC208761 weakly similar to UP|HSF1_ARATH (P41151) Heat shock fac... 572 e-164
TC217188 similar to UP|Q940V6 (Q940V6) Heat shock transcription ... 169 2e-42
TC231490 similar to UP|HSF8_LYCPE (P41153) Heat shock factor pro... 166 2e-41
TC220405 similar to UP|Q43454 (Q43454) Heat shock transcription ... 162 2e-40
TC230981 similar to GB|AAL62005.1|18252249|AY072614 AT3g24520/MO... 160 1e-39
NP005106 heat shock transcription factor 21 152 3e-37
BI471764 similar to SP|Q9SCW5|HSF2_ Heat shock factor protein 2 ... 134 6e-32
TC217234 UP|Q43457 (Q43457) Heat shock transcription factor 34, ... 132 2e-31
BG352891 similar to GP|15810194|gb AT3g24520/MOB24_5 {Arabidopsi... 129 2e-30
TC235016 similar to UP|O23259 (O23259) Heat shock transcription ... 128 5e-30
TC230259 similar to UP|HSF7_ARATH (Q9T0D3) Heat shock factor pro... 118 5e-27
TC206226 similar to UP|HSF6_ARATH (Q9SCW4) Heat shock factor pro... 117 6e-27
TC209126 homologue to UP|Q43456 (Q43456) Heat shock transcriptio... 117 8e-27
TC206227 similar to UP|HSF6_ARATH (Q9SCW4) Heat shock factor pro... 116 1e-26
TC208003 similar to UP|Q6VBB5 (Q6VBB5) Heat shock factor RHSF2, ... 113 1e-25
BM886719 similar to SP|P41152|HSF3_ Heat shock factor protein HS... 108 3e-24
AW620962 101 6e-22
AW132703 96 3e-20
BQ474006 similar to SP|P22335|HSF2_ Heat shock factor protein HS... 91 8e-19
TC219003 homologue to UP|Q43455 (Q43455) Heat shock transcriptio... 87 9e-18
>TC208761 weakly similar to UP|HSF1_ARATH (P41151) Heat shock factor protein
1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1),
partial (24%)
Length = 1285
Score = 572 bits (1475), Expect = e-164
Identities = 283/356 (79%), Positives = 314/356 (87%)
Frame = +2
Query: 11 SSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFI 70
S SV PFL KCYDMV+D +TDS+I W+ G +FVISDIT FSVTLLPTYFKHNNF+SFI
Sbjct: 8 SGSVPPFLKKCYDMVQDCNTDSVICWSH-DGVSFVISDITQFSVTLLPTYFKHNNFSSFI 184
Query: 71 RQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSR 130
RQLNIYGFRK+DTD WEFANENFVRGQKHLLKNI RRKHPH DQQKALP+ +N DEPS+
Sbjct: 185 RQLNIYGFRKIDTDSWEFANENFVRGQKHLLKNIHRRKHPHSADQQKALPQQDNCDEPSQ 364
Query: 131 EAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLSFL 190
EAPNHGL KEVENLK D+NSL QELV L QH ESAE+K+L+LSDRLQGMEKHQQQMLSFL
Sbjct: 365 EAPNHGLWKEVENLKLDKNSLTQELVKLRQHQESAENKLLLLSDRLQGMEKHQQQMLSFL 544
Query: 191 VMVVQSPGFMVQLLHPKENSWRLAEAGNMFDPGKEDDKPVASDGMIVQYKPPVGEKRKHV 250
VMVVQSPGFMVQLLHPKEN+WRLAE+ N D K+DDKPVASDGMI++YKPPVGEK K V
Sbjct: 545 VMVVQSPGFMVQLLHPKENNWRLAESWNRLDQDKQDDKPVASDGMIIKYKPPVGEKLKPV 724
Query: 251 IPIPLSPGFDRQPEPELSADRLKDLCISSEFLKVLLDEKVSPLDNHSPFLLPDLPDDGSW 310
+PLSPGF+ QPEPELSAD LKDLCISSEFLKVLLDEK+SPL+NHSPFLLPDLPDDGSW
Sbjct: 725 --VPLSPGFENQPEPELSADGLKDLCISSEFLKVLLDEKLSPLENHSPFLLPDLPDDGSW 898
Query: 311 EQLFLGSPFLENIEDTDHENEEPTVGGMEMEATIPETPNEHSRIFESMIMEMEKTQ 366
EQLFLGSPFL NI+D+ +E+E T GMEME TI ETPNE+SR FESMI+EM+KT+
Sbjct: 899 EQLFLGSPFLGNIQDSKNESEGHTNSGMEMEPTISETPNENSRTFESMIVEMQKTR 1066
>TC217188 similar to UP|Q940V6 (Q940V6) Heat shock transcription factor,
partial (93%)
Length = 1838
Score = 169 bits (427), Expect = 2e-42
Identities = 96/226 (42%), Positives = 134/226 (58%), Gaps = 4/226 (1%)
Frame = +2
Query: 6 ENASGSSS-VAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHN 64
E A GSSS + PFL K Y+MV+D ST+SI+SW+ + +F++ + FS LLP YFKHN
Sbjct: 533 EEAQGSSSSLPPFLTKTYEMVDDPSTNSIVSWSA-TNRSFIVWNPPEFSRDLLPKYFKHN 709
Query: 65 NFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNN 124
NF+SFIRQLN YGFRK+D ++WEFAN++FVRGQ HLLKNI RRK H Q
Sbjct: 710 NFSSFIRQLNTYGFRKIDPEQWEFANDDFVRGQPHLLKNIHRRKPVHSHSLQNI---QGQ 880
Query: 125 SDEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQ 184
E+ + E+E LK ++ L++EL Q + E ++ +D L+ +EK Q+
Sbjct: 881 GSSLLTESERRSFKDEIEKLKHEKERLLRELERHEQEWKMYEVQLQHSNDCLEKLEKKQE 1060
Query: 185 QMLSFLVMVVQSPGFMVQLLHPKEN---SWRLAEAGNMFDPGKEDD 227
++S + V+Q PG + LL EN RL +G D +D
Sbjct: 1061SLVSSVSQVLQKPGIALNLLLLTENMDRKRRLPRSGLFSDDASIED 1198
>TC231490 similar to UP|HSF8_LYCPE (P41153) Heat shock factor protein HSF8
(Heat shock transcription factor 8) (HSTF 8) (Heat
stress transcription factor), partial (32%)
Length = 646
Score = 166 bits (420), Expect = 2e-41
Identities = 87/163 (53%), Positives = 114/163 (69%), Gaps = 2/163 (1%)
Frame = +3
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
+ +++ PFL+K Y+MVED STDSI+SW+ P+ ++FV+ + F+ LLP +FKHNNF+S
Sbjct: 162 TNANAPPPFLSKTYEMVEDPSTDSIVSWS-PTNNSFVVWNPPEFARDLLPKHFKHNNFSS 338
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEP 128
F+RQLN YGFRKVD DRWEFANE F+RGQKHLLK I RRK H +QQ A H S
Sbjct: 339 FVRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKTITRRKPAHGHNQQ-AQQAHGQSSSV 515
Query: 129 SR--EAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKM 169
E GL +EVE LK D+N LMQELV L Q ++ ++++
Sbjct: 516 GACVEVGKFGLEEEVEILKRDKNVLMQELVRLRQQQQATDNQL 644
>TC220405 similar to UP|Q43454 (Q43454) Heat shock transcription factor 21
(Fragment), partial (83%)
Length = 657
Score = 162 bits (411), Expect = 2e-40
Identities = 82/169 (48%), Positives = 108/169 (63%)
Frame = +1
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K +D+VED ST+ I+SW+ + ++FV+ D FS T+LP YFKHNNF+SF+RQLN
Sbjct: 154 PFLTKTFDVVEDPSTNDIVSWSR-ARNSFVVWDSHKFSTTILPRYFKHNNFSSFVRQLNT 330
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNH 135
YGFRK+D D+WEFANE F+ GQ+ LLK I+RR+H VT Q H E
Sbjct: 331 YGFRKIDPDKWEFANEGFLAGQRQLLKTIKRRRHVTVTQTQ----SHEGGSGACVELGEF 498
Query: 136 GLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQ 184
GL E+E L+ DR LM E+V L Q ++ ++L + RLQ EK Q
Sbjct: 499 GLEGEMERLRRDRTVLMAEIVRLRQQQHNSREQLLSMETRLQATEKKHQ 645
>TC230981 similar to GB|AAL62005.1|18252249|AY072614 AT3g24520/MOB24_5
{Arabidopsis thaliana;} , partial (46%)
Length = 688
Score = 160 bits (404), Expect = 1e-39
Identities = 84/200 (42%), Positives = 125/200 (62%)
Frame = +1
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
+ ++ +APF+ K Y+MV D +TD +I W P+ ++F++ D FS +LLP +FKHNNF+S
Sbjct: 112 NNNNVIAPFVIKTYNMVNDPTTDKLIMWG-PANNSFIVLDPLDFSHSLLPAFFKHNNFSS 288
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEP 128
F+RQLN YGFRKVD DRWEFANE F+RGQKHLL+NI RRKH L H
Sbjct: 289 FVRQLNTYGFRKVDPDRWEFANEWFLRGQKHLLRNIARRKHGGAGRSNFNLHSH------ 450
Query: 129 SREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQMLS 188
+H L+ E + D +++ E+ L + ++ E ++ ++ RL+ EK QQM++
Sbjct: 451 -----HHPLKVE----ELDDEAMVMEIARLKEEQKALEEELQGMNKRLETTEKRPQQMMA 603
Query: 189 FLVMVVQSPGFMVQLLHPKE 208
FL VV+ P + ++L +E
Sbjct: 604 FLSKVVEDPQVLSRILRERE 663
>NP005106 heat shock transcription factor 21
Length = 578
Score = 152 bits (383), Expect = 3e-37
Identities = 83/152 (54%), Positives = 101/152 (65%), Gaps = 1/152 (0%)
Frame = +1
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL+K +DMVED STDSI+SW+ + ++FV+ D FS +LP YFKH NF+SFIRQLN
Sbjct: 133 PFLSKIFDMVEDSSTDSIVSWSM-ARNSFVVWDSHKFSADILPRYFKHGNFSSFIRQLNA 309
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNH 135
YGFRKVD DRWEFANE F+ GQ+HLLK I+RR++ + QQK E
Sbjct: 310 YGFRKVDPDRWEFANEGFLAGQRHLLKTIKRRRNVSQSLQQK------GGSGACVEVGEF 471
Query: 136 GLRKEVENLKSDRNSLMQELVHL-SQHLESAE 166
GL E+E LK DRN LM E+V L Q L S E
Sbjct: 472 GLEGELERLKRDRNILMAEIVRLRHQQLNSRE 567
>BI471764 similar to SP|Q9SCW5|HSF2_ Heat shock factor protein 2 (HSF 2)
(Heat shock transcription factor 2) (HSTF 2). [Mouse-ear
cress], partial (19%)
Length = 421
Score = 134 bits (337), Expect = 6e-32
Identities = 61/99 (61%), Positives = 81/99 (81%)
Frame = +2
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
+GS VAPFL+K YDMV+D STDS++SW + + +TFV+ ++ F+ +LP +FKHNNF+S
Sbjct: 98 NGSVCVAPFLSKTYDMVDDPSTDSVVSWGK-NNNTFVVWNVPQFTTDILPKHFKHNNFSS 274
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRR 107
F+RQLN YGFRKVD DRWEFANE F+RG+K LLK+I R+
Sbjct: 275 FVRQLNTYGFRKVDPDRWEFANEGFLRGEKQLLKSISRQ 391
>TC217234 UP|Q43457 (Q43457) Heat shock transcription factor 34, complete
Length = 1423
Score = 132 bits (332), Expect = 2e-31
Identities = 81/202 (40%), Positives = 103/202 (50%), Gaps = 29/202 (14%)
Frame = +3
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
S+ S S APFL K Y +VED TD +ISW E SG+TFV+ F+ LLP YFKH
Sbjct: 324 SEVEMSQRSVPAPFLTKTYQLVEDQGTDQVISWGE-SGNTFVVWKHADFAKDLLPKYFKH 500
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK-------HP------ 110
NNF+SF+RQLN YGFRK+ D+WEFANE+F RGQK LL I+RRK HP
Sbjct: 501 NNFSSFVRQLNTYGFRKIVPDKWEFANEHFKRGQKELLSEIKRRKTVPQSSAHPPEAGKS 680
Query: 111 ----------------HVTDQQKALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQE 154
+ + N + P+H L E E LK D +L E
Sbjct: 681 GGDGNSPLNSGSDDAGSTSTSSSSSGSKNQGSVETNTTPSHQLSSENEKLKKDNETLSCE 860
Query: 155 LVHLSQHLESAESKMLVLSDRL 176
L++ + + + L DRL
Sbjct: 861 ---LARARKQCDELVAFLRDRL 917
>BG352891 similar to GP|15810194|gb AT3g24520/MOB24_5 {Arabidopsis thaliana},
partial (37%)
Length = 461
Score = 129 bits (325), Expect = 2e-30
Identities = 70/159 (44%), Positives = 102/159 (64%)
Frame = +3
Query: 39 PSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQK 98
P+ ++F++ D FS +LLPT+FKHNNF+SF+RQLN YGFRKVD DRWEFANE F+RGQK
Sbjct: 18 PANNSFIVLDPLDFSYSLLPTFFKHNNFSSFVRQLNTYGFRKVDPDRWEFANEWFLRGQK 197
Query: 99 HLLKNIRRRKHPHVTDQQKALPEHNNSDEPSREAPNHGLRKEVENLKSDRNSLMQELVHL 158
HLL+NI R+KH A ++NS +H L+ E + D +++ E+ L
Sbjct: 198 HLLRNIVRKKH------GGAGRTNSNSHY------SHPLKLE----ELDDEAMVMEIARL 329
Query: 159 SQHLESAESKMLVLSDRLQGMEKHQQQMLSFLVMVVQSP 197
+ ++ E ++ ++ RL+ EK QM++FL VV+ P
Sbjct: 330 KEEQKALEEELHEMNKRLETTEKRPPQMMAFLCKVVEDP 446
>TC235016 similar to UP|O23259 (O23259) Heat shock transcription factor like
protein, partial (12%)
Length = 444
Score = 128 bits (321), Expect = 5e-30
Identities = 60/101 (59%), Positives = 79/101 (77%)
Frame = +2
Query: 8 ASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFA 67
++G+ APFL K YDMV+D ST+ I+SW+ + ++FV+ + F+ LLPTYFKHNNF+
Sbjct: 140 SAGAGGPAPFLLKTYDMVDDASTNDIVSWSS-TNNSFVVWNPPEFARLLLPTYFKHNNFS 316
Query: 68 SFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK 108
SFIRQLN YGFRK+ +RWEFAN+ F++ QKHLLKNI RRK
Sbjct: 317 SFIRQLNTYGFRKIHPERWEFANDEFLKDQKHLLKNIYRRK 439
>TC230259 similar to UP|HSF7_ARATH (Q9T0D3) Heat shock factor protein 7 (HSF
7) (Heat shock transcription factor 7) (HSTF 7), partial
(32%)
Length = 1260
Score = 118 bits (295), Expect = 5e-27
Identities = 58/105 (55%), Positives = 73/105 (69%)
Frame = +1
Query: 4 SKENASGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKH 63
S S S PFL K Y +V+D + D +ISW + SG +F++ + TAF+ LLP YFKH
Sbjct: 100 STSGDSQRSIPTPFLTKTYQLVDDHTIDDVISWND-SGSSFIVWNTTAFAKDLLPKYFKH 276
Query: 64 NNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK 108
NNF+SF+RQLN YGFRKV DRWEF+NE F R +K LL I+RRK
Sbjct: 277 NNFSSFVRQLNTYGFRKVVPDRWEFSNEYFRRDEKRLLCEIQRRK 411
>TC206226 similar to UP|HSF6_ARATH (Q9SCW4) Heat shock factor protein 6 (HSF
6) (Heat shock transcription factor 6) (HSTF 6), partial
(33%)
Length = 535
Score = 117 bits (294), Expect = 6e-27
Identities = 57/100 (57%), Positives = 71/100 (71%)
Frame = +3
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
S S PFL K + +V+D S D +ISW + G TF++ + T F+ LLP YFKHNNF+S
Sbjct: 225 SQRSIPTPFLTKTFQLVDDQSIDDVISWND-DGSTFIVWNPTVFARDLLPKYFKHNNFSS 401
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK 108
F+RQLN YGFRKV DRWEF+NE F RG+K LL I+RRK
Sbjct: 402 FVRQLNTYGFRKVVPDRWEFSNEYFRRGEKRLLCEIQRRK 521
>TC209126 homologue to UP|Q43456 (Q43456) Heat shock transcription factor 31
(Fragment), complete
Length = 1221
Score = 117 bits (293), Expect = 8e-27
Identities = 61/116 (52%), Positives = 77/116 (65%)
Frame = +1
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
S S APFL K Y +V++ +TD I+SW E TFV+ F+ LLP YFKHNNF+S
Sbjct: 112 SHKSVPAPFLTKTYQLVDEPTTDHIVSWGEDDT-TFVVWRPPEFARDLLPNYFKHNNFSS 288
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNN 124
F+RQLN YGFRK+ DRWEFANE F +G+KHLL I RRK QQ ++ H++
Sbjct: 289 FVRQLNTYGFRKIVPDRWEFANEFFKKGEKHLLCEIHRRK--TAQPQQGSMNHHHH 450
>TC206227 similar to UP|HSF6_ARATH (Q9SCW4) Heat shock factor protein 6 (HSF
6) (Heat shock transcription factor 6) (HSTF 6), partial
(54%)
Length = 1489
Score = 116 bits (291), Expect = 1e-26
Identities = 56/100 (56%), Positives = 71/100 (71%)
Frame = +2
Query: 9 SGSSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
S S PFL K Y +V+D S D +ISW + G TF++ + T F+ LLP +FKHNNF+S
Sbjct: 164 SQRSIPTPFLTKTYQLVDDQSIDDVISWND-DGSTFIVWNPTVFARDLLPKFFKHNNFSS 340
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRK 108
F+RQLN YGFRKV DRWEF+N+ F RG+K LL I+RRK
Sbjct: 341 FVRQLNTYGFRKVVPDRWEFSNDYFRRGEKRLLCEIQRRK 460
>TC208003 similar to UP|Q6VBB5 (Q6VBB5) Heat shock factor RHSF2, partial
(15%)
Length = 1105
Score = 113 bits (283), Expect = 1e-25
Identities = 72/189 (38%), Positives = 100/189 (52%), Gaps = 9/189 (4%)
Frame = +2
Query: 16 PFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFIRQLNI 75
PFL K Y +VED +TD +ISW G FV+ F+ LLPT FKH+NF+SF+RQLN
Sbjct: 284 PFLLKTYMLVEDPATDDVISWNA-KGTAFVVWQPPEFARDLLPTLFKHSNFSSFVRQLNT 460
Query: 76 YGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEH---NNSDEPSREA 132
YGFRKV T RWEF N+ F +G++ LL IRRRK + QQ P +SDE R +
Sbjct: 461 YGFRKVATSRWEFFNDKFKKGERELLHEIRRRK-AWTSKQQPNAPNQATLQDSDEDQRSS 637
Query: 133 PNHG------LRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQM 186
L E + LK + L EL + + + + S + ++ ++ M
Sbjct: 638 SISSSSGYTTLVDENKRLKKENGVLNSELTSMKRKCKELLDLVAKYSSHAKEEKEDERPM 817
Query: 187 LSFLVMVVQ 195
L + + VQ
Sbjct: 818 LFGVRLEVQ 844
>BM886719 similar to SP|P41152|HSF3_ Heat shock factor protein HSF30 (Heat
shock transcription factor 30) (HSTF 30), partial (18%)
Length = 422
Score = 108 bits (271), Expect = 3e-24
Identities = 59/140 (42%), Positives = 84/140 (59%)
Frame = +3
Query: 66 FASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNS 125
F+SF+RQLN YGFRKVD+DRWEFANE F G+KHLLKNIRRR ++N
Sbjct: 3 FSSFVRQLNTYGFRKVDSDRWEFANEGFQGGKKHLLKNIRRR------------CKYNKL 146
Query: 126 DEPSREAPNHGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRLQGMEKHQQQ 185
+ + + EVE LK D+N L E++ L Q E++ ++ + +R++ E Q Q
Sbjct: 147 HQGAFNMMKPCVDSEVEKLKKDQNILKVEILKLRQQQENSHVQLTNVQERIRCAEVKQYQ 326
Query: 186 MLSFLVMVVQSPGFMVQLLH 205
M+ FL + + P F+ QL+H
Sbjct: 327 MMYFLTRMARRPAFVEQLVH 386
>AW620962
Length = 337
Score = 101 bits (251), Expect = 6e-22
Identities = 56/111 (50%), Positives = 70/111 (62%)
Frame = +2
Query: 11 SSSVAPFLNKCYDMVEDDSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFASFI 70
S SV PFL KCYDMV+D +TDS I + G +F +SD T ++HN+ + +
Sbjct: 8 SGSVPPFLKKCYDMVQDCNTDSPILGSHHGG-SFAMSDQTQSLRRPFSPSWQHNHCSRVL 184
Query: 71 RQLNIYGFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPE 121
RQL+IYGFRK+DTD EF ENFV G LL +I RRK P D QKALP+
Sbjct: 185 RQLHIYGFRKIDTDSREFGYENFVMGPNPLLNSIHRRKPPPSADLQKALPQ 337
>AW132703
Length = 420
Score = 95.5 bits (236), Expect = 3e-20
Identities = 49/102 (48%), Positives = 64/102 (62%), Gaps = 2/102 (1%)
Frame = +2
Query: 77 GFRKVDTDRWEFANENFVRGQKHLLKNIRRRKHPHVTDQQKALPEHNNSDEPSR--EAPN 134
GFRKVD DRWEFANE F++GQKHLL++I RRK H + Q+ H S E
Sbjct: 8 GFRKVDPDRWEFANEGFLKGQKHLLRSITRRKPAHGQNHQQPQQPHGQSSSVGACVEVGK 187
Query: 135 HGLRKEVENLKSDRNSLMQELVHLSQHLESAESKMLVLSDRL 176
GL +EVE LK D+N LMQELV L Q ++ ++++ + RL
Sbjct: 188 FGLEEEVERLKRDKNVLMQELVRLRQQQQTTDNQLQTMVQRL 313
>BQ474006 similar to SP|P22335|HSF2_ Heat shock factor protein HSF24 (Heat
shock transcription factor 24) (HSTF 24), partial (23%)
Length = 421
Score = 90.9 bits (224), Expect = 8e-19
Identities = 47/93 (50%), Positives = 61/93 (65%), Gaps = 6/93 (6%)
Frame = +1
Query: 15 APFLNKCYDMVED------DSTDSIISWTEPSGHTFVISDITAFSVTLLPTYFKHNNFAS 68
APFL K Y+++E+ DST I+SW G FV+ FS LP YFKHNNF+S
Sbjct: 148 APFLLKTYELLEELSENEKDST-KIVSWNA-EGTGFVVWSPAEFSELTLPRYFKHNNFSS 321
Query: 69 FIRQLNIYGFRKVDTDRWEFANENFVRGQKHLL 101
FIRQLN YGF+K+ + +WEF +E F RG +H+L
Sbjct: 322 FIRQLNTYGFKKISSKKWEFKHEKFQRGCRHIL 420
>TC219003 homologue to UP|Q43455 (Q43455) Heat shock transcription factor 29
(Fragment), partial (76%)
Length = 1188
Score = 87.4 bits (215), Expect = 9e-18
Identities = 40/65 (61%), Positives = 51/65 (77%)
Frame = +1
Query: 44 FVISDITAFSVTLLPTYFKHNNFASFIRQLNIYGFRKVDTDRWEFANENFVRGQKHLLKN 103
F++ F+ LLP YFKHNNF+SF+RQLN YGFRKV DRWEFAN+ F RG++ LL++
Sbjct: 10 FIVWRPAEFARDLLPKYFKHNNFSSFVRQLNTYGFRKVVPDRWEFANDCFRRGERALLRD 189
Query: 104 IRRRK 108
I+RRK
Sbjct: 190 IQRRK 204
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,361,694
Number of Sequences: 63676
Number of extensions: 240563
Number of successful extensions: 1121
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 1093
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1097
length of query: 367
length of database: 12,639,632
effective HSP length: 99
effective length of query: 268
effective length of database: 6,335,708
effective search space: 1697969744
effective search space used: 1697969744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0069.1


