

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
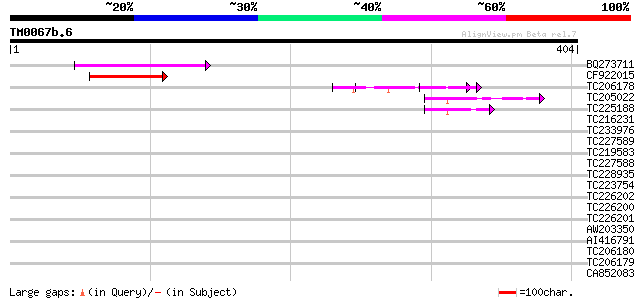
Query= TM0067b.6
(404 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BQ273711 86 4e-17
CF922015 61 8e-10
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 46 3e-05
TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, pa... 43 2e-04
TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like prot... 42 5e-04
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 40 0.002
TC233976 40 0.002
TC227589 40 0.002
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 39 0.003
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 39 0.003
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 39 0.004
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 39 0.006
TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.009
TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.009
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 38 0.009
AW203350 37 0.016
AI416791 37 0.016
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 37 0.016
TC206179 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 36 0.027
CA852083 34 0.10
>BQ273711
Length = 409
Score = 85.5 bits (210), Expect = 4e-17
Identities = 41/97 (42%), Positives = 58/97 (59%)
Frame = -2
Query: 47 RQREITLDQNKGLNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATY 106
++R++ + +GL FR+ +PPKFSG DP+ A LW+ E EKIF + E KV AT+
Sbjct: 291 QERDVEPAEYRGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAMGCLEEHKVLYATF 112
Query: 107 LLLGDAEYWWKGTRGIMEDNHEEISWNSFRTAFLEKY 143
+L G+AE WWK + I WN+F+ FLE Y
Sbjct: 111 MLQGEAENWWKFVKPSFVAPGGVIPWNAFKDKFLENY 1
>CF922015
Length = 172
Score = 61.2 bits (147), Expect = 8e-10
Identities = 26/55 (47%), Positives = 39/55 (70%)
Frame = -3
Query: 58 GLNDFRRQNPPKFSGGTDPDKADLWIQEVEKIFGVLQTAEGAKVRMATYLLLGDA 112
GL+ F+R NPP F GG DP+ A+ W++E+EKIF V++ + KV AT++L +A
Sbjct: 167 GLDRFQRNNPPTFKGGYDPEGAEAWLREIEKIFRVMECQDHQKVLFATHMLADEA 3
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 45.8 bits (107), Expect = 3e-05
Identities = 18/44 (40%), Positives = 26/44 (58%)
Frame = +2
Query: 293 ETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPKTEP 336
++ C+ C EPGH A++CP+ C C + GH A +C AP P
Sbjct: 449 KSLCWNCKEPGHMASSCPN-EGICHTCGKAGHRARECSAPPMPP 577
Score = 43.9 bits (102), Expect = 1e-04
Identities = 25/86 (29%), Positives = 36/86 (41%), Gaps = 3/86 (3%)
Frame = +2
Query: 247 SQNFQSREKFQSRGPYQRPAGT---GFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPG 303
S + +SR + +SR P R + + YR + ++ C C PG
Sbjct: 209 SSDSRSRSRSRSRSPMDRKIRSDRFSYRDAPYRR-------DSRRGFSRDNLCKNCKRPG 367
Query: 304 HYANACPDTRPKCFNCNRMGHTAGQC 329
HYA CP+ C NC GH A +C
Sbjct: 368 HYARECPNV-AICHNCGLPGHIASEC 442
Score = 42.4 bits (98), Expect = 4e-04
Identities = 29/101 (28%), Positives = 43/101 (41%), Gaps = 2/101 (1%)
Frame = +2
Query: 231 NRRLNRAGTGGPM--RSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQ 288
+R +R+ + PM + S F R+ PY+R + GF+ + G +
Sbjct: 218 SRSRSRSRSRSPMDRKIRSDRFSYRD-----APYRRDSRRGFSRDNLCKNCKRPGHYARE 382
Query: 289 TLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
C CG PGH A+ C T+ C+NC GH A C
Sbjct: 383 C-PNVAICHNCGLPGHIASECT-TKSLCWNCKEPGHMASSC 499
Score = 37.0 bits (84), Expect = 0.016
Identities = 17/37 (45%), Positives = 19/37 (50%)
Frame = +2
Query: 293 ETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
E C C + GH A CP+ P C CN GH A QC
Sbjct: 641 EKACNNCRKTGHLARDCPND-PICNLCNVSGHVARQC 748
Score = 35.4 bits (80), Expect = 0.047
Identities = 17/50 (34%), Positives = 22/50 (44%)
Frame = +2
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
GA GG G ++ C C + GH + C C NC GH A +C
Sbjct: 800 GARGGGGGGY--RDVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYEC 943
Score = 35.4 bits (80), Expect = 0.047
Identities = 17/47 (36%), Positives = 20/47 (42%), Gaps = 6/47 (12%)
Frame = +2
Query: 293 ETTCFRCGEPGHYANAC------PDTRPKCFNCNRMGHTAGQCRAPK 333
E C CG+ GH A C P C NC + GH A +C K
Sbjct: 506 EGICHTCGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEK 646
>TC205022 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (69%)
Length = 1623
Score = 43.1 bits (100), Expect = 2e-04
Identities = 30/89 (33%), Positives = 41/89 (45%), Gaps = 3/89 (3%)
Frame = +2
Query: 296 CFRCGEPGHYANACP--DTRPKCFNCNRMGHTAGQCR-APKTEPTVNTARGKRPAAKARV 352
CF CG GH+A C D + KC+ C GH C+ +PK ++T RG R
Sbjct: 314 CFNCGLDGHWARDCKAGDWKNKCYRCGERGHIERNCKNSPK---KLSTRRG-------RS 463
Query: 353 YTMDGERAEEFARGERKNDGNFLTILSYS 381
Y+ R+ RG R D ++ SYS
Sbjct: 464 YSRSPVRSHSPRRG-RSRDRSYSRDRSYS 547
>TC225188 similar to UP|Q9FYB7 (Q9FYB7) Splicing factor-like protein, partial
(62%)
Length = 1131
Score = 42.0 bits (97), Expect = 5e-04
Identities = 20/52 (38%), Positives = 25/52 (47%), Gaps = 2/52 (3%)
Frame = +1
Query: 296 CFRCGEPGHYANACP--DTRPKCFNCNRMGHTAGQCRAPKTEPTVNTARGKR 345
CF CG GH+A C D + KC+ C GH C K P + RG+R
Sbjct: 364 CFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNC---KNSPKKLSQRGRR 510
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 40.0 bits (92), Expect = 0.002
Identities = 32/107 (29%), Positives = 36/107 (32%), Gaps = 13/107 (12%)
Frame = +3
Query: 236 RAGTGGPMRSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETT 295
R G GG R G RG Y G G+ G Y G GG G
Sbjct: 102 RGGYGGGGRGG------------RGGYGGGGG-GYGGGGYGGGGGYGGGGGGGG----GG 230
Query: 296 CFRCGEPGHYANACPDTRP-------------KCFNCNRMGHTAGQC 329
C++CGE GH A C C+NC GH A C
Sbjct: 231 CYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDC 371
Score = 33.1 bits (74), Expect = 0.23
Identities = 22/75 (29%), Positives = 29/75 (38%)
Frame = +3
Query: 238 GTGGPMRSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCF 297
G GG + G +R+ Q G R G G GG G +C+
Sbjct: 219 GGGGCYKCGETGHIARDCSQGGGGGGRYGGGG-------------GGGG--------SCY 335
Query: 298 RCGEPGHYANACPDT 312
CGE GH+A CP +
Sbjct: 336 NCGESGHFARDCPSS 380
>TC233976
Length = 763
Score = 39.7 bits (91), Expect = 0.002
Identities = 29/123 (23%), Positives = 49/123 (39%), Gaps = 8/123 (6%)
Frame = +1
Query: 221 EKATEVELMKNRRLNRAGTGGPM-RSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMT 279
++ +V KN + PM S ++ + + ++++ QR +G +G +
Sbjct: 22 DQQEKVAFYKNANASHGKEKKPMTHSRAKPYSAPLEYENHYGGQRTSGGHHLAGGSSQLV 201
Query: 280 GAA------GGSGDQTL-KKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAP 332
GGSG + C +CG GH A+ C CFN GH + C P
Sbjct: 202 NRVSQPAGRGGSGAPAIVTTPLRCRKCGRLGHNAHECTYREVTCFNYQGKGHLSTNCPHP 381
Query: 333 KTE 335
+ E
Sbjct: 382 RKE 390
>TC227589
Length = 547
Score = 39.7 bits (91), Expect = 0.002
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Frame = +2
Query: 296 CFRCGEPGHYANACPDTRPK--CFNCNRMGHTAGQC 329
CF CGE GH A C + K C+ C +GH A QC
Sbjct: 47 CFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQC 154
Score = 38.5 bits (88), Expect = 0.006
Identities = 19/55 (34%), Positives = 25/55 (44%), Gaps = 9/55 (16%)
Frame = +2
Query: 287 DQTLKKETTCFRCGEPGHYANAC---PD------TRPKCFNCNRMGHTAGQCRAP 332
D E +C++CG+ GH AC PD T C C GH A +C +P
Sbjct: 377 DDATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSSCCKCGEAGHFAQECTSP 541
Score = 35.8 bits (81), Expect = 0.036
Identities = 17/51 (33%), Positives = 24/51 (46%)
Frame = +2
Query: 291 KKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPKTEPTVNTA 341
K++ C+ CG GH A C + CF C + GH A C T + + A
Sbjct: 98 KRKKPCYVCGGLGHNARQCTKAQD-CFICKKGGHRAKDCLEKHTSRSKSVA 247
Score = 28.9 bits (63), Expect = 4.4
Identities = 14/41 (34%), Positives = 19/41 (46%), Gaps = 7/41 (17%)
Frame = +2
Query: 291 KKETTCFRCGEPGHYANAC-----PD--TRPKCFNCNRMGH 324
K C +CG GH +C PD +C+ C R+GH
Sbjct: 236 KSVAICLKCGNSGHDMFSCRNDYSPDDLKEIQCYVCKRVGH 358
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 39.3 bits (90), Expect = 0.003
Identities = 20/54 (37%), Positives = 23/54 (42%)
Frame = +2
Query: 258 SRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACPD 311
+R Y G G G R G GG G CF CGE GH+A CP+
Sbjct: 455 ARDCYHGQGGGGGDDGRNRRRGGGGGGGGG--------CFNCGEEGHFARECPN 592
Score = 32.3 bits (72), Expect = 0.39
Identities = 25/83 (30%), Positives = 29/83 (34%), Gaps = 20/83 (24%)
Frame = +2
Query: 267 GTGFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANAC---------PDTRPK-- 315
G G G Y G G G + E C+ CG GH A C D R +
Sbjct: 344 GRGRGGGRYGGGEGRGRGFGRRGGGPE--CYNCGRIGHLARDCYHGQGGGGGDDGRNRRR 517
Query: 316 ---------CFNCNRMGHTAGQC 329
CFNC GH A +C
Sbjct: 518 GGGGGGGGGCFNCGEEGHFAREC 586
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 39.3 bits (90), Expect = 0.003
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Frame = +1
Query: 296 CFRCGEPGHYANACPDTRPK--CFNCNRMGHTAGQC 329
CF CGE GH A C + K C+ C +GH A QC
Sbjct: 61 CFNCGEEGHAAVNCSAVKRKKPCYVCGCLGHNARQC 168
Score = 37.4 bits (85), Expect = 0.012
Identities = 17/52 (32%), Positives = 23/52 (43%), Gaps = 9/52 (17%)
Frame = +1
Query: 287 DQTLKKETTCFRCGEPGHYANACPDTRPK---------CFNCNRMGHTAGQC 329
D E +C++CG+ GH AC R + CF C GH A +C
Sbjct: 391 DDATPGEISCYKCGQLGHTGLACSRLRDEITSGATPSSCFKCGEEGHFAREC 546
Score = 35.4 bits (80), Expect = 0.047
Identities = 17/52 (32%), Positives = 25/52 (47%)
Frame = +1
Query: 290 LKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPKTEPTVNTA 341
+K++ C+ CG GH A C + CF C + GH A C T + + A
Sbjct: 109 VKRKKPCYVCGCLGHNARQCSKVQD-CFICKKGGHRAKDCPEKHTSTSKSIA 261
Score = 32.7 bits (73), Expect = 0.30
Identities = 16/54 (29%), Positives = 19/54 (34%), Gaps = 7/54 (12%)
Frame = +1
Query: 284 GSGDQTLKKETTCFRCGEPGHYANACPDTRPK-------CFNCNRMGHTAGQCR 330
G + K CF C + GH A CP+ C C GH CR
Sbjct: 148 GHNARQCSKVQDCFICKKGGHRAKDCPEKHTSTSKSIAICLKCGNSGHDIFSCR 309
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 38.9 bits (89), Expect = 0.004
Identities = 18/51 (35%), Positives = 21/51 (40%), Gaps = 6/51 (11%)
Frame = +1
Query: 291 KKETTCFRCGEPGHYANACPDT------RPKCFNCNRMGHTAGQCRAPKTE 335
+K C RC GH A CP+ C+NC GH QC P E
Sbjct: 331 EKNKICLRCRRRGHRAKNCPEVLDGAKDAKYCYNCGENGHALTQCLHPLQE 483
Score = 32.3 bits (72), Expect = 0.39
Identities = 15/45 (33%), Positives = 18/45 (39%), Gaps = 5/45 (11%)
Frame = +1
Query: 290 LKKETTCFRCGEPGHYANACP-----DTRPKCFNCNRMGHTAGQC 329
+K +CF C H A CP + C C R GH A C
Sbjct: 253 MKPGESCFICKAMDHIAKLCPEKAEWEKNKICLRCRRRGHRAKNC 387
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 38.5 bits (88), Expect = 0.006
Identities = 31/108 (28%), Positives = 35/108 (31%), Gaps = 9/108 (8%)
Frame = +2
Query: 231 NRRLNRAGTGGPMRSGSQNFQSREKFQSRGPYQRPAGTGFTSGSYRPMTGAAGGSGDQTL 290
NR N G GG G N G + A G G GG G
Sbjct: 86 NRSSNSGGGGGGSGGGCFNC---------GGFGHLARDCVRGGGGSVGIGGGGGGG---- 226
Query: 291 KKETTCFRCGEPGHYANACPDTRPK---------CFNCNRMGHTAGQC 329
+CFRCG GH A C + CF C +GH A C
Sbjct: 227 ----SCFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDC 358
Score = 38.1 bits (87), Expect = 0.007
Identities = 22/62 (35%), Positives = 25/62 (39%), Gaps = 15/62 (24%)
Frame = +2
Query: 283 GGSGDQTLKKETTCFRCGEPGHYANAC---------------PDTRPKCFNCNRMGHTAG 327
GGSG CFRCGE GH A C + CFNC + GH A
Sbjct: 299 GGSGGG-------CFRCGEVGHLARDCGMEGGRFGGGGGSGGGGGKSTCFNCGKPGHFAR 457
Query: 328 QC 329
+C
Sbjct: 458 EC 463
Score = 37.7 bits (86), Expect = 0.009
Identities = 16/41 (39%), Positives = 22/41 (53%)
Frame = +2
Query: 269 GFTSGSYRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
G G + G+ GG G ++TCF CG+PGH+A C
Sbjct: 359 GMEGGRFGGGGGSGGGGG------KSTCFNCGKPGHFAREC 463
Score = 34.7 bits (78), Expect = 0.079
Identities = 16/44 (36%), Positives = 19/44 (42%), Gaps = 10/44 (22%)
Frame = +2
Query: 296 CFRCGEPGHYANACPDTRPK----------CFNCNRMGHTAGQC 329
C++CGE GH A C + CFNC GH A C
Sbjct: 44 CYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDC 175
Score = 32.0 bits (71), Expect = 0.51
Identities = 21/66 (31%), Positives = 21/66 (31%), Gaps = 12/66 (18%)
Frame = +2
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANACP------------DTRPKCFNCNRMGHTAG 327
G GGSG CF CG GH A C CF C GH A
Sbjct: 107 GGGGGSGGG-------CFNCGGFGHLARDCVRGGGGSVGIGGGGGGGSCFRCGGFGHMAR 265
Query: 328 QCRAPK 333
C K
Sbjct: 266 DCATGK 283
>TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 1003
Score = 37.7 bits (86), Expect = 0.009
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = +3
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
G GG G ++ + C+ CGEPGH+A C
Sbjct: 786 GGGGGRGGRSGGSDLKCYECGEPGHFAREC 875
>TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (89%)
Length = 936
Score = 37.7 bits (86), Expect = 0.009
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = +2
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
G GG G ++ + C+ CGEPGH+A C
Sbjct: 290 GGGGGRGGRSGGSDLKCYECGEPGHFAREC 379
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 37.7 bits (86), Expect = 0.009
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = +1
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANAC 309
G GG G ++ + C+ CGEPGH+A C
Sbjct: 199 GGGGGRGGRSGGSDLKCYECGEPGHFAREC 288
>AW203350
Length = 411
Score = 37.0 bits (84), Expect = 0.016
Identities = 21/61 (34%), Positives = 30/61 (48%)
Frame = +1
Query: 294 TTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQCRAPKTEPTVNTARGKRPAAKARVY 353
T C++CG+PGH++ CP + P N N +TA T P NT + K+R
Sbjct: 10 TGCYKCGKPGHWSRDCPFSAP---NPNPNSNTA-------TTPNPNTPNPSSSSFKSRSA 159
Query: 354 T 354
T
Sbjct: 160 T 162
>AI416791
Length = 420
Score = 37.0 bits (84), Expect = 0.016
Identities = 37/127 (29%), Positives = 55/127 (43%), Gaps = 7/127 (5%)
Frame = -3
Query: 128 EEISWNSFRTAFLEKYFPTSARDERESQFLTLRQGGMSVPEFASKLESLAKHFQFFHDHV 187
E+++W S + A LE+Y D E Q L+Q G SV ++ ++ E L +
Sbjct: 385 EDLTWESLKIALLERYGGHGDGDVYE-QLTELKQEG-SVEDYITEFEYLIAQI----PRL 224
Query: 188 NERYMCKRFVNGLRPDIEDSVRPLRIMRFQS-----LVEKATEVELM--KNRRLNRAGTG 240
E+ F++GL+ +I VR L M S V +A E E+ R
Sbjct: 223 PEKQFQGYFLHGLQSEIRGRVRTLATMGEMSRTKLLQVTRAVEKEVKGGNGSGFGRGPKN 44
Query: 241 GPMRSGS 247
GP RS S
Sbjct: 43 GPYRSNS 23
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 37.0 bits (84), Expect = 0.016
Identities = 17/37 (45%), Positives = 19/37 (50%)
Frame = +3
Query: 293 ETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
E C C + GH A CP+ P C CN GH A QC
Sbjct: 126 EKACNNCRKTGHLARDCPND-PICNLCNVSGHVARQC 233
Score = 35.4 bits (80), Expect = 0.047
Identities = 17/50 (34%), Positives = 22/50 (44%)
Frame = +3
Query: 280 GAAGGSGDQTLKKETTCFRCGEPGHYANACPDTRPKCFNCNRMGHTAGQC 329
GA GG G ++ C C + GH + C C NC GH A +C
Sbjct: 276 GARGGGGGGY--RDVVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYEC 419
Score = 32.0 bits (71), Expect = 0.51
Identities = 15/41 (36%), Positives = 18/41 (43%), Gaps = 6/41 (14%)
Frame = +3
Query: 299 CGEPGHYANAC------PDTRPKCFNCNRMGHTAGQCRAPK 333
CG+ GH A C P C NC + GH A +C K
Sbjct: 9 CGKAGHRARECSAPPMPPGDLRLCNNCYKQGHIAAECTNEK 131
>TC206179 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (16%)
Length = 407
Score = 36.2 bits (82), Expect = 0.027
Identities = 27/91 (29%), Positives = 35/91 (37%), Gaps = 3/91 (3%)
Frame = +3
Query: 241 GPMRSGSQNFQSREKFQSRGPYQRPAGT---GFTSGSYRPMTGAAGGSGDQTLKKETTCF 297
G + + SR + +SR P R + + YR G S D K
Sbjct: 159 GKKKKLKMSSDSRSRSRSRSPMDRKIRSDRFSYRDAPYR-RDSRRGFSRDNLGKN----- 320
Query: 298 RCGEPGHYANACPDTRPKCFNCNRMGHTAGQ 328
C PGHYA CP+ C NC GH A +
Sbjct: 321 -CKRPGHYARECPNV-AICHNCGLPGHIASE 407
>CA852083
Length = 829
Score = 34.3 bits (77), Expect = 0.10
Identities = 32/115 (27%), Positives = 47/115 (40%), Gaps = 4/115 (3%)
Frame = +1
Query: 202 PDIEDSVRPLRIMRFQSLVEKATEV-ELMKNRRLNRAGTGGPMRSGSQNFQSREKFQSRG 260
PDI+D R L L T + E K+ + + AG S E+ ++R
Sbjct: 109 PDIDDDFRELYKEYTGPLGTATTNMQERAKSNKRSNAG-------------SDEEEEARD 249
Query: 261 PYQRPAGTGFTSGS---YRPMTGAAGGSGDQTLKKETTCFRCGEPGHYANACPDT 312
P P T FTS + + A + + ++E C CGE GH+ CP T
Sbjct: 250 PNAVP--TDFTSREAKVWEAKSKATERNWKKRKEEEMICKLCGESGHFTQGCPST 408
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,829,820
Number of Sequences: 63676
Number of extensions: 212737
Number of successful extensions: 1339
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 1238
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1308
length of query: 404
length of database: 12,639,632
effective HSP length: 99
effective length of query: 305
effective length of database: 6,335,708
effective search space: 1932390940
effective search space used: 1932390940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0067b.6


