

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0066.5
(667 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
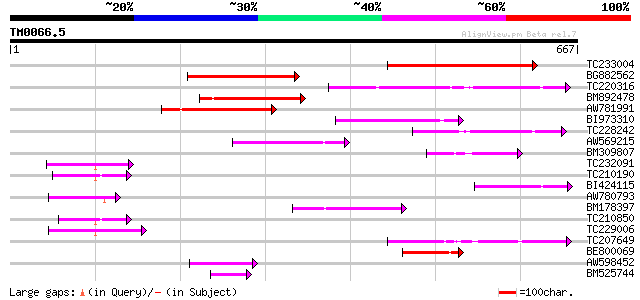
Score E
Sequences producing significant alignments: (bits) Value
TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] ... 214 8e-56
BG882562 200 1e-51
TC220316 154 1e-37
BM892478 137 2e-32
AW781991 123 3e-28
BI973310 119 6e-27
TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired r... 99 5e-21
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 75 7e-14
BM309807 similar to GP|5764395|gb| far-red impaired response pro... 75 1e-13
TC232091 69 7e-12
TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 64 2e-10
BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidop... 62 8e-10
AW780793 62 1e-09
BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thalia... 61 2e-09
TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, part... 61 2e-09
TC229006 60 4e-09
TC207649 similar to UP|Q9SY66 (Q9SY66) F14N23.12 (At1g10240/F14N... 57 2e-08
BE800069 47 2e-05
AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {A... 45 1e-04
BM525744 44 3e-04
>TC233004 weakly similar to PIR|G96565|G96565 F6D8.26 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(8%)
Length = 551
Score = 214 bits (546), Expect = 8e-56
Identities = 97/177 (54%), Positives = 134/177 (74%)
Frame = +1
Query: 445 YVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFM 504
YVHK T KEF+DKY+L LH+KH +E++AD E+RN S LKTRC+ E+QL+++YT+EIF
Sbjct: 7 YVHKHTSFKEFVDKYDLVLHRKHLKEAMADLETRNVSFELKTRCNFEVQLAKVYTKEIFQ 186
Query: 505 KFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCIC 564
KFQ EVE M+SCF T Q+ V+G I+ Y+VKERV ++GN + +K FEV+Y ++RCIC
Sbjct: 187 KFQSEVEGMYSCFNTRQVSVNGSIITYVVKERVEVEGNEKGVKSFEVLYETTELDIRCIC 366
Query: 565 SCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPI 621
S FN+ GYLCR AL VLN+NG+EEIP +YIL RW++D+K++ H + D +P+
Sbjct: 367 SLFNYKGYLCRRALNVLNYNGIEEIPSRYILHRWRRDFKQMFNQFHVYDNVDSHNPV 537
>BG882562
Length = 416
Score = 200 bits (509), Expect = 1e-51
Identities = 90/131 (68%), Positives = 109/131 (82%)
Frame = +3
Query: 210 RGDTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTY 269
+GD + I N+ CR+QL NPNFFY+MD +D+G LRN FW D+RSRAA YFGDV+ FD+T
Sbjct: 24 KGDPELISNYFCRIQLMNPNFFYVMDLNDDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTC 203
Query: 270 LSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDR 329
LS+ YEIPLV FVG+NHHG+SVLLGCGLLA ET E+YIWLFR W+TCM+G PQTIIT+
Sbjct: 204 LSNNYEIPLVAFVGVNHHGKSVLLGCGLLADETFETYIWLFRAWLTCMTGRPPQTIITNX 383
Query: 330 CEALQSAIIEV 340
C+A+QSAI EV
Sbjct: 384 CKAMQSAIAEV 416
>TC220316
Length = 988
Score = 154 bits (389), Expect = 1e-37
Identities = 88/286 (30%), Positives = 149/286 (51%), Gaps = 2/286 (0%)
Frame = +1
Query: 376 KAVYETLKISEFEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPG 435
K Y++ + E A W ++G+ D WL+ +Y+ R W P++LK FF G+
Sbjct: 1 KVCYQSQTVDEXXATWNXXXNKYGLKDDAWLKEMYQKRASWVPLYLKGTFFAGIPM---N 171
Query: 436 ESIGPFFGRYVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLS 495
ES+ FFG ++ QTPL EF+ +YE L ++ +EE D + N P+L+T+ +E Q
Sbjct: 172 ESLDSFFGALLNAQTPLMEFIPRYERGLERRREEERKEDFNTSNFQPILQTKEPVEEQCR 351
Query: 496 RMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSR 555
R+YT +F FQ E+ + FS G ++ +G + Y+V+ GN +++ V ++
Sbjct: 352 RLYTLTVFKIFQKELLQCFSYLG-FKIFEEGGLSRYMVRR----CGN--DMEKHVVTFNA 510
Query: 556 AAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSA 615
+ + C C F + G LCRH L V + E+P +YIL RW ++ + PD S S+
Sbjct: 511 SNISISCSCQMFEYEGVLCRHVLRVFQILQLREVPSRYILHRWTRNTEDGVFPDMESWSS 690
Query: 616 DD--TDPIQWSNQLFSSALQVVEEGTISLDHYNVALQACEESLSKV 659
+ + WS L +A + ++ G S++ Y +A + E K+
Sbjct: 691 SQELKNLMLWS--LRETASKYIDAGATSIEKYKLAYEILREGGRKL 822
>BM892478
Length = 429
Score = 137 bits (345), Expect = 2e-32
Identities = 63/125 (50%), Positives = 84/125 (66%)
Frame = +3
Query: 224 QLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVG 283
Q + FFY M+ D+ G+ N FWAD RSR +C +FGDV+ D +Y + Y +P FVG
Sbjct: 54 QAEDTGFFYAMEVDN-GNCMNIFWADGRSRYSCSHFGDVLVLDTSYRKTVYLVPFATFVG 230
Query: 284 INHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSPQTIITDRCEALQSAIIEVFPK 343
+NHH Q VLLGC L+A E+ ES+ WLF+TW+ MSG P T+I D+ A+Q AI +VFP
Sbjct: 231 VNHHKQPVLLGCALIADESEESFTWLFQTWLRAMSGRLPLTVIADQDIAIQRAIAKVFPV 410
Query: 344 SHHCF 348
+HH F
Sbjct: 411 THHRF 425
>AW781991
Length = 419
Score = 123 bits (308), Expect = 3e-28
Identities = 54/135 (40%), Positives = 81/135 (60%)
Frame = +3
Query: 179 LVIDAGGNGNLNPSARDDRTFSEMSNKLNVKRGDTQAIYNFLCRMQLTNPNFFYLMDFDD 238
L+ + GG + + D R + +N+L GD Q + ++L +M NPNFFY + D+
Sbjct: 18 LIKEYGGISKVGFTEVDCRNYMR-NNRLRSLEGDIQLVLDYLRQMHAENPNFFYAVQGDE 194
Query: 239 EGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLL 298
+ + N FWAD ++R +FGD + FD TY S++Y +P F G+NHHGQ VL GC L
Sbjct: 195 DQSITNVFWADPKARMNYTFFGDTVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGCAFL 374
Query: 299 AGETTESYIWLFRTW 313
E+ S++WLF+TW
Sbjct: 375 INESEASFVWLFKTW 419
>BI973310
Length = 457
Score = 119 bits (297), Expect = 6e-27
Identities = 63/152 (41%), Positives = 90/152 (58%), Gaps = 2/152 (1%)
Frame = +3
Query: 384 ISEFEAAWGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFG 443
I EFE+ W +++RF V D+EWL+S+Y R W PV+L++ FFG +S E + FF
Sbjct: 6 IDEFESYWHSLLERFYVMDNEWLQSIYNSRQHWVPVYLRETFFGEISLNEGNEYLISFFD 185
Query: 444 RYVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIF 503
YV T L+ + +YE A+ H++E AD ++ NSSPVLKT +E Q + +YTR+IF
Sbjct: 186 GYVFSSTTLQVLVRQYEKAVSSWHEKELKADYDTTNSSPVLKTPSPMEKQAASLYTRKIF 365
Query: 504 MKFQFEVEEMFSCFGTTQLHVD--GPIVIYLV 533
MKFQ EE+ + +D G I Y V
Sbjct: 366 MKFQ---EELVETLANPAIKIDDSGTITTYRV 452
>TC228242 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein; Mutator-like transposase-like protein;
phytochrome A signaling protein-like, partial (25%)
Length = 1354
Score = 99.4 bits (246), Expect = 5e-21
Identities = 64/182 (35%), Positives = 90/182 (49%)
Frame = +2
Query: 474 DTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLV 533
D+++ N LKT LE ++ ++T +F K Q EV +C D IV
Sbjct: 2 DSDTWNKVATLKTPSPLEKSVAGIFTHAVFKKIQAEVIGAVACHPKADRQDDTTIV---- 169
Query: 534 KERVLIDGNRREIKDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKY 593
RV + KDF VV ++ EL CIC F + GYLCRHAL VL ++G P +Y
Sbjct: 170 -HRV---HDMETNKDFFVVVNQVKSELSCICRLFEYRGYLCRHALFVLQYSGQSVFPSQY 337
Query: 594 ILSRWKKDYKRLHVPDHSSVSADDTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQACE 653
IL RW KD K ++ S +Q N L AL++ EEG++S + Y +A A
Sbjct: 338 ILKRWTKDAKVRNIMGEESEHM--LTRVQRYNDLCQRALKLSEEGSLSQESYGIAFHALH 511
Query: 654 ES 655
E+
Sbjct: 512 EA 517
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 75.5 bits (184), Expect = 7e-14
Identities = 41/139 (29%), Positives = 72/139 (51%), Gaps = 2/139 (1%)
Frame = +1
Query: 263 IYFDNTYLSSKYEIPLVVFVGINHHGQSVLLGCGLLAGETTESYIWLFRTWVTCMSGCSP 322
+ FD TY Y++ L +++G++++G + C LL E +S+ W + ++ M G +P
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKAP 189
Query: 323 QTIITDRCEALQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGL--HNYDGIRKALIKAVYE 380
QTI+TD L+ AI P++ H F + I+ K + L YD KA +Y
Sbjct: 190 QTILTDHNMWLKEAIAVELPQTKHAFCIWHILSKFSDWFSLLLGSQYDE-WKAEFHRLYN 366
Query: 381 TLKISEFEAAWGIMIQRFG 399
++ +FE W M+ ++G
Sbjct: 367 LEQVEDFEEGWRQMVDQYG 423
>BM309807 similar to GP|5764395|gb| far-red impaired response protein
{Arabidopsis thaliana}, partial (15%)
Length = 428
Score = 74.7 bits (182), Expect = 1e-13
Identities = 42/113 (37%), Positives = 60/113 (52%)
Frame = +3
Query: 491 ELQLSRMYTREIFMKFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFE 550
E Q+S +YT IF KFQ EV + C + DG I ++V++ + ++F
Sbjct: 48 EKQMSTVYTHAIFKKFQVEVLGVAGCQSRIEAG-DGTIAKFIVQDY-------EKDEEFL 203
Query: 551 VVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYK 603
V ++ + E+ C C F + G+LCRHAL VL G +P YIL RW KD K
Sbjct: 204 VTWNELSSEVSCFCRLFEYKGFLCRHALSVLQRCGCSCVPSHYILKRWTKDAK 362
>TC232091
Length = 530
Score = 68.9 bits (167), Expect = 7e-12
Identities = 36/107 (33%), Positives = 56/107 (51%), Gaps = 5/107 (4%)
Frame = +1
Query: 44 DAPAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGF---- 99
+ P VGMEF+S + A N+Y YA+ GF VR+ ++ C+ QGF
Sbjct: 130 EEPNVGMEFQSEEAAKNFYEEYARREGFVVRLDRCHRSEVDKQIISRRFSCNKQGFHVRV 309
Query: 100 -KRTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGA 145
+TK V+ R R GC AMM +++ +W + + + EH+H+L A
Sbjct: 310 RNKTKPVHKPRASIREGCEAMMYVKVNTCGKWVVTKFVKEHSHLLNA 450
>TC210190 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (65%)
Length = 866
Score = 64.3 bits (155), Expect = 2e-10
Identities = 39/97 (40%), Positives = 49/97 (50%), Gaps = 4/97 (4%)
Frame = +1
Query: 51 EFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGF----KRTKDVN 106
EF S A+ +Y YA EVGF VRV R G L C+ +GF KR K V
Sbjct: 4 EFGSEAAAHAFYNAYATEVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFRMADKREKIVR 183
Query: 107 HLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
R ETR GC AM+ +R S +W + + + EH H L
Sbjct: 184 Q-RAETRVGCRAMIMVRKLSSGKWVVAKFVKEHTHPL 291
>BI424115 similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (9%)
Length = 402
Score = 62.0 bits (149), Expect = 8e-10
Identities = 35/118 (29%), Positives = 61/118 (51%), Gaps = 2/118 (1%)
Frame = +2
Query: 547 KDFEVVYSRAAGELRCICSCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKR-L 605
K + V + + + C C F + G LC+H L V V +P YIL RW ++ K
Sbjct: 44 KAYMVTLNHSELKANCSCQMFEYAGILCKHILTVFTVTNVLTLPPHYILKRWTRNAKNSA 223
Query: 606 HVPDHSSVS-ADDTDPIQWSNQLFSSALQVVEEGTISLDHYNVALQACEESLSKVHDV 662
+ +H+ S A ++ ++ N L A++ EEG+++++ YN A+ E + KV +V
Sbjct: 224 GLDEHTGESHAQESLTARYGN-LCKEAIRYAEEGSVTVETYNAAISGLREGVKKVANV 394
>AW780793
Length = 419
Score = 61.6 bits (148), Expect = 1e-09
Identities = 29/91 (31%), Positives = 50/91 (54%), Gaps = 6/91 (6%)
Frame = +3
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGFKRTKDV 105
P +GMEF S ++A +YI Y + +GF VR+ ++ R + + G CS +GF+ K +
Sbjct: 132 PFIGMEFNSREEAREFYIAYGRRIGFTVRIHHNRRSRVNNQVIGQDFVCSKEGFRAKKYL 311
Query: 106 NHLRK------ETRTGCPAMMRIRIAESQRW 130
+ + TR GC AM+R+ + + +W
Sbjct: 312 HRKDRVLPPPPATREGCQAMIRLALRDRGKW 404
>BM178397 similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana}, partial
(20%)
Length = 422
Score = 60.8 bits (146), Expect = 2e-09
Identities = 38/136 (27%), Positives = 63/136 (45%), Gaps = 2/136 (1%)
Frame = +1
Query: 333 LQSAIIEVFPKSHHCFGLSLIMKKVPEKLGGL--HNYDGIRKALIKAVYETLKISEFEAA 390
L+ A+ P + H F + +I+ K P + Y+ KA +Y + +FE
Sbjct: 13 LKEALSTEMPTTKHAFCIWMIVAKFPSWFNAVLGERYNDW-KAEFYRLYNLESVEDFELG 189
Query: 391 WGIMIQRFGVSDHEWLRSLYEDRVRWAPVFLKDVFFGGMSAARPGESIGPFFGRYVHKQT 450
W M FG+ + + +LY R WA FL+ F GM+ +SI F R++ QT
Sbjct: 190 WREMACSFGLHSNRHMVNLYSSRSLWALPFLRSHFLAGMTTTGQSKSINAFIQRFLSAQT 369
Query: 451 PLKEFLDKYELALHKK 466
L F+++ +A+ K
Sbjct: 370 RLAHFVEQVAVAVDFK 417
>TC210850 similar to UP|Q93Z68 (Q93Z68) AT3g59470/T16L24_20, partial (63%)
Length = 1326
Score = 60.8 bits (146), Expect = 2e-09
Identities = 36/90 (40%), Positives = 46/90 (51%), Gaps = 4/90 (4%)
Frame = +1
Query: 58 AYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGF----KRTKDVNHLRKETR 113
A+ +Y YA +VGF VRV R G L C+ +GF KR K V R ETR
Sbjct: 7 AHAFYNAYATDVGFIVRVSKLSRSRRDGTAIGRTLVCNKEGFRMADKREKIVRQ-RAETR 183
Query: 114 TGCPAMMRIRIAESQRWRIIEVILEHNHIL 143
GC AM+ +R S +W I + + EH H L
Sbjct: 184 VGCRAMIMVRKLSSGKWVIAKFVKEHTHPL 273
>TC229006
Length = 991
Score = 59.7 bits (143), Expect = 4e-09
Identities = 37/120 (30%), Positives = 57/120 (46%), Gaps = 5/120 (4%)
Frame = +2
Query: 46 PAVGMEFESYDDAYNYYICYAKEVGFCVRVKNSWFKRNSREKYGAVLCCSSQGF-----K 100
P GMEFES D A +Y YA+ +GF +RV + L C+ +G+
Sbjct: 101 PYEGMEFESEDAAKIFYDEYARRLGFVMRVMSCRRPERDGRILARRLGCNKEGYCVSIRG 280
Query: 101 RTKDVNHLRKETRTGCPAMMRIRIAESQRWRIIEVILEHNHILGAKIHKSAKKMGNGTKR 160
+ V R TR GC AM+ I+ +S +W I + + +HNH L ++ + M K+
Sbjct: 281 KFASVRKPRASTREGCKAMIHIKYNKSGKWVITKFVKDHNHPLVVSPREARQTMDEKDKK 460
>TC207649 similar to UP|Q9SY66 (Q9SY66) F14N23.12 (At1g10240/F14N23_12),
partial (30%)
Length = 971
Score = 57.4 bits (137), Expect = 2e-08
Identities = 48/217 (22%), Positives = 97/217 (44%)
Frame = +2
Query: 445 YVHKQTPLKEFLDKYELALHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFM 504
++ QT L F+++ +A+ K + + + LKT +E + + T F
Sbjct: 116 FLSAQTRLAHFVEQVAVAVDFKDQTG*QQTMQQNLQNVCLKTGAPMESHAATILTPFAFS 295
Query: 505 KFQFEVEEMFSCFGTTQLHVDGPIVIYLVKERVLIDGNRREIKDFEVVYSRAAGELRCIC 564
K Q ++ + + + + + DG +LV+ +G R+ V ++ G + C C
Sbjct: 296 KLQEQLV-LAAHYASFSIE-DG----FLVRHHTKAEGGRK------VYWAPQEGIISCSC 439
Query: 565 SCFNFYGYLCRHALCVLNFNGVEEIPFKYILSRWKKDYKRLHVPDHSSVSADDTDPIQWS 624
F F G LCRH+L VL+ +IP +Y+ RW +R+++P + + D +
Sbjct: 440 HQFEFTGILCRHSLRVLSTGNCFQIPDRYLPIRW----RRINMPSSKLLQSAPNDHAERV 607
Query: 625 NQLFSSALQVVEEGTISLDHYNVALQACEESLSKVHD 661
L + ++ E S + ++A + LS++ +
Sbjct: 608 KLLQNMVSSLMTESAKSKERLDIATEQVTLLLSRIRE 718
>BE800069
Length = 233
Score = 47.4 bits (111), Expect = 2e-05
Identities = 25/71 (35%), Positives = 43/71 (60%)
Frame = +1
Query: 463 LHKKHKEESLADTESRNSSPVLKTRCSLELQLSRMYTREIFMKFQFEVEEMFSCFGTTQL 522
L ++++E AD ++ N+ PVL+T +E Q S +YTR+IFM+FQ E+ + ++
Sbjct: 1 LESRNEKEVRADYDTMNTLPVLRTPSPMEKQASELYTRKIFMRFQEELVGTLTLM-ASKA 177
Query: 523 HVDGPIVIYLV 533
DG ++ Y V
Sbjct: 178 DDDGEVITYHV 210
>AW598452 weakly similar to GP|15982769|gb| At2g27110/T20P8.16 {Arabidopsis
thaliana}, partial (7%)
Length = 256
Score = 45.1 bits (105), Expect = 1e-04
Identities = 22/80 (27%), Positives = 35/80 (43%)
Frame = +2
Query: 212 DTQAIYNFLCRMQLTNPNFFYLMDFDDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLS 271
D + + RMQ N FY + D++ + N A RSR + + GD D Y
Sbjct: 17 DAHNLLEYCKRMQAENHGCFYAIQLDEDDRVSNVL*AGTRSRTSYKHMGDAESLDAAYGK 196
Query: 272 SKYEIPLVVFVGINHHGQSV 291
+Y + + G N HG+ +
Sbjct: 197 HRYAVSYAPYTGRNCHGRMI 256
>BM525744
Length = 411
Score = 43.5 bits (101), Expect = 3e-04
Identities = 18/48 (37%), Positives = 29/48 (59%)
Frame = +3
Query: 237 DDEGHLRNAFWADARSRAACGYFGDVIYFDNTYLSSKYEIPLVVFVGI 284
DD +R+ FWA + G F V++ DNTY ++Y++PL+ VG+
Sbjct: 210 DDSDAIRDIFWAHPDAIKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGV 353
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,020,449
Number of Sequences: 63676
Number of extensions: 500633
Number of successful extensions: 2481
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 2456
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2472
length of query: 667
length of database: 12,639,632
effective HSP length: 104
effective length of query: 563
effective length of database: 6,017,328
effective search space: 3387755664
effective search space used: 3387755664
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0066.5


