

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.18
(799 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
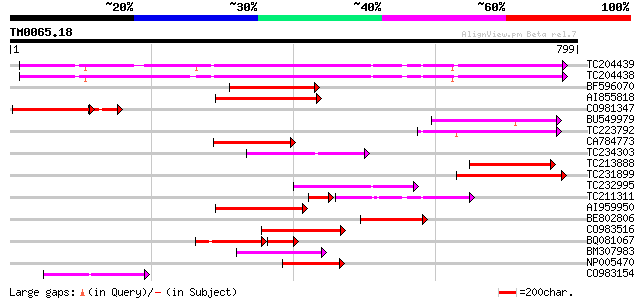
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 467 e-132
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 464 e-131
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 143 3e-34
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 141 1e-33
CO981347 111 2e-30
BU549979 125 7e-29
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 125 1e-28
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 122 6e-28
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 120 2e-27
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 114 2e-25
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 113 4e-25
TC232995 110 2e-24
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 91 3e-23
AI959950 106 5e-23
BE802806 105 6e-23
CO983516 102 5e-22
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 77 6e-22
BM307983 100 3e-21
NP005470 reverse transcriptase 100 3e-21
CO983154 92 7e-19
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 467 bits (1202), Expect = e-132
Identities = 270/787 (34%), Positives = 427/787 (53%), Gaps = 15/787 (1%)
Frame = +1
Query: 14 IKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNERARCMRIQS 73
IK ++SD+G E+++ F +FC+ GI + TP+QNG+ ER NRTL E AR M
Sbjct: 2443 IKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAK 2622
Query: 74 GLPKMFWVDAINTAAYLINRGPSIPLDYQLPE---EVWSGKEVSLSHLKVFGCVSYVLID 130
LP W +A+NTA Y+ NR + L P E+W G++ S+ H +FG Y+L D
Sbjct: 2623 ELPYNLWAEAMNTACYIHNR---VTLRRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILAD 2793
Query: 131 SDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDRSSAESMS 190
++R K+DPK+ F+GY + YR ++ + + ++ S NV ++ +S
Sbjct: 2794 REQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDD------------LS 2937
Query: 191 SSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIETPITQVRRSTR 250
+++ + E V + +D K + N EN D + +P T I ++
Sbjct: 2938 PARKKDVEEDVRTSGDNVADAAKSGE-NAENSDSATDESNINQPDKRSSTRIQKMHPKEL 3114
Query: 251 TPKAPQRYSPS-------LNYLLLTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQKNG 303
P R + ++ EP+ EA+ W AM++E+ ++N
Sbjct: 3115 IIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTDEF---WINAMQEELEQFKRNE 3285
Query: 304 TWSLTKLPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQQKQGIDFTEIFSPVVKM 362
W L PEG + +W+++ K +G R KARLV +G+ Q +G+DF E F+PV ++
Sbjct: 3286 VWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARL 3465
Query: 363 TTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLY 422
+IR++L + L Q+DVK AFL+G L EE+Y+ QP+GF + V +L K+LY
Sbjct: 3466 ESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKALY 3645
Query: 423 GLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVDDMLIAGSNMTEI 482
GLKQAPR WY++ EF++ G+ + +D FVK+ A++ +I +YVDD++ G + +
Sbjct: 3646 GLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEML 3825
Query: 483 NRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRS 542
QQM FEM +G LG+++ + E + LSQ +Y + ++ +F + +A+ +
Sbjct: 3826 RHFVQQMQSEFEMSLVGELTYFLGLQV--KQMEDSIFLSQSRYAKNIVKKFGMENASHKR 3999
Query: 543 TPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSN 602
TP HLK SK DE + + Y S +GSL+Y + +RPDI +AVGV +R+ +N
Sbjct: 4000 TPAPTHLKLSK------DEAGTSVDQSLYRSMIGSLLY-LTASRPDITYAVGVCARYQAN 4158
Query: 603 PGKEHWECVKWILRYLKGSS----RMCLCFRRNNLTLQEFSDADLGGDSDGGKSTTGYIF 658
P H VK IL+Y+ G+S C C +N L + DAD G +D KST+G F
Sbjct: 4159 PKISHLTQVKRILKYVNGTSDYGIMYCHC---SNPMLVGYCDADWAGSADDRKSTSGGCF 4329
Query: 659 TLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQ 718
LG +SW SK QN V+LST E+EY+A + +++W+K LKE EQDV L+ D+
Sbjct: 4330 YLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNM 4509
Query: 719 SVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRL 778
S I ++KNPV HSR KHI +++H+IR+L+ D+ ++L + E D+ TK + A+
Sbjct: 4510 SAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQFEK 4689
Query: 779 CIASAGL 785
G+
Sbjct: 4690 LRGKLGI 4710
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 464 bits (1194), Expect = e-131
Identities = 266/781 (34%), Positives = 424/781 (54%), Gaps = 9/781 (1%)
Frame = +1
Query: 14 IKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNERARCMRIQS 73
IK ++SD+G E+++ +F +FC+ GI + TP+QNG+ ER NRTL E AR M
Sbjct: 2446 IKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAK 2625
Query: 74 GLPKMFWVDAINTAAYLINRGPSIPLDYQLPE---EVWSGKEVSLSHLKVFGCVSYVLID 130
LP W +A+NTA Y+ NR + L P E+W G++ ++ H +FG Y+L D
Sbjct: 2626 ELPYNLWAEAMNTACYIHNR---VTLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILAD 2796
Query: 131 SDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDRSSAESMS 190
++R K+DPK+ F+GY + YR ++ + + ++ S NV ++ + + E +
Sbjct: 2797 REQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPARKKDVEEDVR 2976
Query: 191 SSKQLKLSERVALEEISESD-VVKRNQINPENEDDEVEVELEQEPTTVIETPITQVRRST 249
+S + E SD IN ++ + ++ +I P V +
Sbjct: 2977 TSGDNVADTAKSAENAENSDSATDEPNINQPDKRPSIRIQKMHPKELIIGDPNRGVTTRS 3156
Query: 250 RTPKAPQRYSPSLNYLLLTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTK 309
R + ++ EP+ EA+ W AM++E+ ++N W L
Sbjct: 3157 REIEI-------VSNSCFVSKIEPKNVKEALTDEF---WINAMQEELEQFKRNEVWELVP 3306
Query: 310 LPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVI 368
PEG + +W+++ K +G R KARLV +G+ Q +G+DF E F+PV ++ +IR++
Sbjct: 3307 RPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLL 3486
Query: 369 LSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAP 428
L + L Q+DVK AFL+G L EE Y+ QP+GF + V +L K+LYGLKQAP
Sbjct: 3487 LGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAP 3666
Query: 429 RQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQ 488
R WY++ EF++ G+ + +D FVK+ A++ +I +YVDD++ G + + QQ
Sbjct: 3667 RAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQ 3846
Query: 489 MSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNH 548
M FEM +G LG+++ + E + LSQ KY + ++ +F + +A+ + TP H
Sbjct: 3847 MQSEFEMSLVGELTYFLGLQV--KQMEDSIFLSQSKYAKNIVKKFGMENASHKRTPAPTH 4020
Query: 549 LKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHW 608
LK SK DE + + Y S +GSL+Y + +RPDI +AVGV +R+ +NP H
Sbjct: 4021 LKLSK------DEAGTSVDQSLYRSMIGSLLY-LTASRPDITYAVGVCARYQANPKISHL 4179
Query: 609 ECVKWILRYLKGSS----RMCLCFRRNNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTA 664
VK IL+Y+ G+S C C ++ L + DAD G +D KST+G F LG
Sbjct: 4180 NQVKRILKYVNGTSDYGIMYCHC---SDSMLVGYCDADWAGSADDRKSTSGGCFYLGTNL 4350
Query: 665 VSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLA 724
+SW SK QN V+LST E+EY+A + +++W+K LKE EQDV L+ D+ S I ++
Sbjct: 4351 ISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINIS 4530
Query: 725 KNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRLCIASAG 784
KNPV HSR KHI +++H+IR+L+ D+ ++L + E D+ TK + A+ G
Sbjct: 4531 KNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKALDANQFEKLRGKLG 4710
Query: 785 L 785
+
Sbjct: 4711 I 4713
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 143 bits (360), Expect = 3e-34
Identities = 72/128 (56%), Positives = 89/128 (69%), Gaps = 1/128 (0%)
Frame = -2
Query: 310 LPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVI 368
LP GK + RWVY +K G R KARLV KG+ Q GID+ + FSPV K+TT+R+
Sbjct: 400 LPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVRLF 221
Query: 369 LSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAP 428
L++ A + L QLD+K AFLHGDLEE+IYM QP GF G LVCKLH+SLYGLKQ+P
Sbjct: 220 LAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQSP 41
Query: 429 RQWYKKFN 436
R W+ KF+
Sbjct: 40 RAWFGKFS 17
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 141 bits (356), Expect = 1e-33
Identities = 66/151 (43%), Positives = 101/151 (66%), Gaps = 1/151 (0%)
Frame = -3
Query: 290 LAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQKQ 348
+AM++E+ ++N W L + PE + +WV+R K + G R KARLV KG+ Q++
Sbjct: 461 IAMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEE 282
Query: 349 GIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVL 408
GID+ E ++PV ++ IR++L+ V+ N L Q+DVK AFL+G ++EE+Y+ QP GFE+
Sbjct: 281 GIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIP 102
Query: 409 GTKNLVCKLHKSLYGLKQAPRQWYKKFNEFM 439
V KL K+LYGLKQAPR WY++ + F+
Sbjct: 101 DKPTHVYKLQKALYGLKQAPRAWYERISNFL 9
>CO981347
Length = 624
Score = 111 bits (277), Expect(2) = 2e-30
Identities = 55/116 (47%), Positives = 76/116 (65%)
Frame = +2
Query: 4 TEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLN 63
T + NQ K+K L++DNG E+ ++F +FC + GI+ K + TP QNG+AERMN T+
Sbjct: 131 TLIGNQLGTKLKVLRTDNGLEFVLEQFNEFCRKIGIKRHKIVPHTP*QNGLAERMNMTIL 310
Query: 64 ERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLK 119
ER RCM + + LPK FW +A NT +YLINR PS L ++ P E WSG+ L +K
Sbjct: 311 ERVRCMLLSARLPKTFWGEAANTTSYLINRCPSSTLGFKTPMEAWSGETT*LFRIK 478
Score = 40.4 bits (93), Expect(2) = 2e-30
Identities = 20/49 (40%), Positives = 30/49 (60%)
Frame = +3
Query: 111 KEVSLSHLKVFGCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFW 159
K + S LKVFG +++ D ++ KLD +A+KC FIGY + Y+ W
Sbjct: 453 KPPNYSGLKVFGSLAF---DHVKQGKLDARAVKCVFIGYPKGVKRYKLW 590
>BU549979
Length = 615
Score = 125 bits (314), Expect = 7e-29
Identities = 68/187 (36%), Positives = 106/187 (56%), Gaps = 4/187 (2%)
Frame = -1
Query: 595 VVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNN-LTLQEFSDADLGGDSDGGKST 653
V+ R+ SNPG +HW+ K ++RYL+G+ L +++ N L + +SD+D G D +ST
Sbjct: 615 VLGRYQSNPGIDHWKTAKKVMRYLQGTKDYMLMYKQTNCLEVIGYSDSDFAGCVDSRRST 436
Query: 654 TGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVP-- 711
+GYIF L VSW+S Q +A ST E E+V EA +WLKSF+ L +
Sbjct: 435 SGYIFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDSISRP 256
Query: 712 -PLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKT 770
L+ D+ + +F+AKN +R KHI +KY IRE + ++++ + + D LTK
Sbjct: 255 LKLYCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTELMIVDPLTKG 76
Query: 771 VTADNLR 777
+T N +
Sbjct: 75 MTPKNFK 55
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (7%)
Length = 804
Score = 125 bits (313), Expect = 1e-28
Identities = 72/208 (34%), Positives = 117/208 (55%), Gaps = 5/208 (2%)
Frame = +1
Query: 575 VGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF----RR 630
+GSL Y + +RP+I AV ++SRFM P H + K +LR +KG+ + F +
Sbjct: 34 IGSLRY-LCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGVLFPFKAKS 210
Query: 631 NNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEA 690
L ++D+D D + KST GY+F V+ SK Q+ +ALST E+EYVA S
Sbjct: 211 GKPDLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCEAEYVAASLG 390
Query: 691 AKEMIWLKSFLKELG-KEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISD 749
A + +W+ + L+EL +E+ L D++S I LAK+P H R KHI++++H+IR+ +S
Sbjct: 391 ACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRFHYIRDQVSK 570
Query: 750 EELSLLKILGSENPTDMLTKTVTADNLR 777
+++ E D++TK + +
Sbjct: 571 GNVTVEYCKAEEQLADLMTKPIQVSRFK 654
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 122 bits (306), Expect = 6e-28
Identities = 61/116 (52%), Positives = 78/116 (66%), Gaps = 1/116 (0%)
Frame = +3
Query: 288 WELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQQ 346
W AM DEM +L+ NGTW L LP GK + RWVY +K +G R KARLV KG+ Q
Sbjct: 57 WRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNGKVDRLKARLVAKGYTQ 236
Query: 347 KQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQP 402
GI++ + FSPV +TT+R+ L++ A + L QLD+K AFLHGDLEE+IYM QP
Sbjct: 237 VYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLHGDLEEDIYMEQP 404
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 120 bits (301), Expect = 2e-27
Identities = 73/175 (41%), Positives = 99/175 (55%), Gaps = 1/175 (0%)
Frame = +1
Query: 334 RYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDL 393
++KARLV K + Q G D+T FSPV KM + ++ S+ + L LD K AFLHG L
Sbjct: 40 QFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFLHGYL 219
Query: 394 EEEIYMTQPEGFEVLG-TKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHC 452
EEE+YM QP GF G + N+VC+L +S YGLKQ+PR W F + ++ + DH
Sbjct: 220 EEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAW--PFLYCGAAIWYDSHEADHS 393
Query: 453 CFVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGM 507
F I L +YVDD+ I GS+ I LK + F+ KDLG + LG+
Sbjct: 394 VFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 114 bits (284), Expect = 2e-25
Identities = 57/123 (46%), Positives = 78/123 (63%), Gaps = 1/123 (0%)
Frame = +3
Query: 648 DGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKE 707
D KSTTG++F +G TA +W SK Q V LST E+EYVA + IWL++ LKEL
Sbjct: 9 DDRKSTTGFVFFMGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELKMP 188
Query: 708 QDVP-PLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDM 766
Q+ P + D++S + LAKNPVFH + KHI +YHFIRE I +E+ L ++ + D+
Sbjct: 189 QEEPMEICVDNKSALALAKNPVFHEKSKHIDTRYHFIRECIEKKEVKLKYVMSQDQAADI 368
Query: 767 LTK 769
TK
Sbjct: 369 FTK 377
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (30%)
Length = 687
Score = 113 bits (282), Expect = 4e-25
Identities = 58/156 (37%), Positives = 94/156 (60%), Gaps = 1/156 (0%)
Frame = +2
Query: 630 RNNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISE 689
+ N L + DAD G +ST+GY +GG VSWKSK Q VA S+ E+EY +++
Sbjct: 2 KGNTQLSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAM 181
Query: 690 AAKEMIWLKSFLKELGKEQDVP-PLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELIS 748
E++W+K FL+EL +++ L+ D+Q+ + +A NPVFH R KHI++ HFIRE +
Sbjct: 182 VTCELMWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLL 361
Query: 749 DEELSLLKILGSENPTDMLTKTVTADNLRLCIASAG 784
+E+ I ++ P D+LTK++ +++ + G
Sbjct: 362 SKEIVTEFIGSNDQPVDILTKSLRGPKIQIVCSKLG 469
>TC232995
Length = 1009
Score = 110 bits (275), Expect = 2e-24
Identities = 56/176 (31%), Positives = 96/176 (53%)
Frame = +2
Query: 401 QPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFAD 460
QP GFE+ N V KL K+LYGLKQAPR WY++ + F+ F+R +D F+K+ +
Sbjct: 8 QPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRKHN 187
Query: 461 SYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKL 520
+++ +YVDD++ +N + M FEM +G K LG++I +++ + +
Sbjct: 188 DILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQI--KQTQ*GIFI 361
Query: 521 SQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVG 576
+Q KY ++L+ RF + A STP+ + K +S Q+ + + Y + +G
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVVEIG 529
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 90.5 bits (223), Expect(2) = 3e-23
Identities = 61/196 (31%), Positives = 100/196 (50%), Gaps = 1/196 (0%)
Frame = +3
Query: 460 DSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLK 519
++++I+ +YVDD++ ++ + M + FE G K +LG++I + + G+
Sbjct: 480 ETFLIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQ-KVYGIF- 653
Query: 520 LSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLM 579
+ QEKY + L RF + +A +TP+ K DE+ ++ S Y+ + SL
Sbjct: 654 IHQEKYTKSHLKRFRMDEAKPMATPMHRSTIIDK------DEKGNHTS*KEYSGMIDSLS 815
Query: 580 YAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNNLTLQEF 638
Y + +RPDI V + +RF S P H VK ILRYL G++ CL F +R+ L +
Sbjct: 816 Y-LTSSRPDIVFVVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGY 992
Query: 639 SDADLGGDSDGGKSTT 654
D GD KST+
Sbjct: 993 CDVYFAGDKVERKSTS 1040
Score = 37.0 bits (84), Expect(2) = 3e-23
Identities = 16/36 (44%), Positives = 23/36 (63%)
Frame = +2
Query: 421 LYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVK 456
+YGLKQA R WY++ + F+ ++GF R D F K
Sbjct: 362 VYGLKQALRAWYERLSSFLVSNGFTRGITDPALFRK 469
>AI959950
Length = 466
Score = 106 bits (264), Expect = 5e-23
Identities = 55/130 (42%), Positives = 83/130 (63%), Gaps = 1/130 (0%)
Frame = -1
Query: 291 AMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQKQG 349
AM++E+ QKN L KLP+ KK + +W++ K + DG RYKARLV KG+ Q++G
Sbjct: 391 AMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEG 212
Query: 350 IDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLG 409
ID+ + F+ V ++ I ++LS N+ L Q+DVK AFL+G +++E+Y+ QP GFE
Sbjct: 211 IDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFENET 32
Query: 410 TKNLVCKLHK 419
V KL+K
Sbjct: 31 LHQHVFKLNK 2
>BE802806
Length = 285
Score = 105 bits (263), Expect = 6e-23
Identities = 50/94 (53%), Positives = 70/94 (74%)
Frame = -2
Query: 495 MKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKK 554
MKDLG A++ILG+ I R+R++G L LSQ Y++K+++RF + + STPLG+H K S
Sbjct: 284 MKDLGSARRILGIDIHRDRAKGELFLSQSNYLKKVVERFRMHQSKPVSTPLGHHTKLSVT 105
Query: 555 QSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPD 588
Q+P+T EE S M+ PYA+ VGS+MY MVC+RPD
Sbjct: 104 QAPETAEERSKMNQTPYANGVGSIMYGMVCSRPD 3
>CO983516
Length = 724
Score = 102 bits (255), Expect = 5e-22
Identities = 49/118 (41%), Positives = 77/118 (64%)
Frame = +2
Query: 356 FSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVC 415
F PV ++ +IR++L + L Q+DVK AFL+G L EE+Y+ QP+GF + V
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 416 KLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVDDML 473
+L K+LYGLKQAPR WY++ E ++ G+ + +D FVK+ A++ +I +YVDD++
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIV 718
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 76.6 bits (187), Expect(2) = 6e-22
Identities = 42/102 (41%), Positives = 62/102 (60%), Gaps = 1/102 (0%)
Frame = +1
Query: 262 LNYLLLTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRW 321
L+ LL EP +A+ S W AM+ + +L +N T +LT LP GK A+ +W
Sbjct: 1 LHPTLLFTTAEPSTVKQALI---SPPWRQAMQADFDALMENKTLTLTSLPSGKAAIDCKW 171
Query: 322 VYRLKEESDGS-RRYKARLVVKGFQQKQGIDFTEIFSPVVKM 362
V+R+KE G+ RY++RLV KGF K G D++E FSPV+++
Sbjct: 172 VFRIKENLYGTLNRYRSRLVAKGFHLKFGCDYSETFSPVIEL 297
Score = 46.6 bits (109), Expect(2) = 6e-22
Identities = 23/43 (53%), Positives = 29/43 (66%)
Frame = +3
Query: 364 TIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFE 406
TIR+IL I + L+Q+D+ AFLHG L EE+YM Q GFE
Sbjct: 300 TIRLILFIALTNHWPLQQVDINNAFLHGLLTEEVYMVQLPGFE 428
>BM307983
Length = 406
Score = 100 bits (249), Expect = 3e-21
Identities = 56/129 (43%), Positives = 76/129 (58%), Gaps = 2/129 (1%)
Frame = +2
Query: 320 RWVYRLKEESDGSR-RYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAE-NL 377
RW+Y +K +D + RYKARLV KG+ Q GID+ E F+ K A+
Sbjct: 11 RWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQFGW 190
Query: 378 HLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNE 437
+ Q DVK AFLHG LEEE+YM P G+ N VC+L K+LYGLKQ+PR W+ +F +
Sbjct: 191 EMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGRFTQ 370
Query: 438 FMSNSGFNR 446
M + G+ +
Sbjct: 371 AMLSLGYKQ 397
>NP005470 reverse transcriptase
Length = 267
Score = 100 bits (248), Expect = 3e-21
Identities = 50/89 (56%), Positives = 60/89 (67%), Gaps = 1/89 (1%)
Frame = +1
Query: 385 KIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGF 444
+ AFLHG LEE I M QPEGFEV G + V +L +SLYGLKQ+PRQWY F+ F++N GF
Sbjct: 1 RTAFLHGRLEENILMKQPEGFEVQGKERYVSQLQRSLYGLKQSPRQWYMSFDSFITNQGF 180
Query: 445 NRCDMDHCCFVKKFADSYII-LALYVDDM 472
R D C + K D +I L LYVDDM
Sbjct: 181 KRSLYDCCVYHNKVEDGLMIYLLLYVDDM 267
>CO983154
Length = 568
Score = 92.4 bits (228), Expect = 7e-19
Identities = 56/152 (36%), Positives = 82/152 (53%), Gaps = 2/152 (1%)
Frame = +3
Query: 48 TPEQNGVAERMNRTLNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEV 107
TP+QNG+AER NR L E AR + + +P W DA+ T+ +LINR PS L+ Q+P +
Sbjct: 6 TPQQNGIAERKNRHLLETARSLMLNLNVPIHHWGDAVLTSCFLINRMPSSSLENQIPHSL 185
Query: 108 WSGKEVSLSHL--KVFGCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKK 165
+ L H+ KVFGC +V S DKL +++KC F+GY GY+ + ++
Sbjct: 186 VFPHD-PLFHVSPKVFGCTCFVHDLSPGLDKLSARSVKCVFLGYSRLQKGYKCYSPTMRR 362
Query: 166 IIRSRNVTFNESVLYKDRSSAESMSSSKQLKL 197
S +VTF E + S S S + L +
Sbjct: 363 YYMSADVTFFEDTPFFSPSVDHSSSLQEVLPI 458
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,598,167
Number of Sequences: 63676
Number of extensions: 431429
Number of successful extensions: 2161
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 2098
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2110
length of query: 799
length of database: 12,639,632
effective HSP length: 105
effective length of query: 694
effective length of database: 5,953,652
effective search space: 4131834488
effective search space used: 4131834488
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0065.18


