

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.12
(1309 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
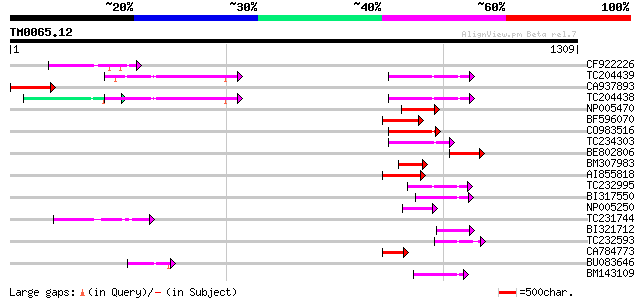
Score E
Sequences producing significant alignments: (bits) Value
CF922226 149 8e-36
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 121 2e-27
CA937893 similar to GP|20805072|dbj retrovirus-related pol polyp... 120 4e-27
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 119 1e-26
NP005470 reverse transcriptase 114 4e-25
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 94 4e-19
CO983516 90 7e-18
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 88 2e-17
BE802806 88 3e-17
BM307983 82 1e-15
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 82 2e-15
TC232995 79 2e-14
BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Ara... 71 4e-12
NP005250 reverse transcriptase 65 3e-10
TC231744 64 6e-10
BI321712 63 1e-09
TC232593 weakly similar to UP|Q9XG91 (Q9XG91) Tpv2-1c protein (F... 60 5e-09
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 58 2e-08
BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {V... 56 1e-07
BM143109 55 2e-07
>CF922226
Length = 667
Score = 149 bits (376), Expect = 8e-36
Identities = 87/228 (38%), Positives = 127/228 (55%), Gaps = 13/228 (5%)
Frame = -3
Query: 89 MSKTLMNKLFAKQRLYSLKMQEGGDLQAHVYAFNNILADLTRLRVKVDDEDKAIILLRSL 148
M+K+L+N+L+ KQ LYS KM E + + FN ++ DL + V +DDED+A++LL L
Sbjct: 665 MTKSLVNRLYXKQSLYSFKMHEDRSVGEQLDLFNKLILDLENIDVTIDDEDQALLLLCYL 486
Query: 149 PGSYDHLVTTLTYGKDSIKLDSISSTLLQHAQRRQSVEEGGGSSGKCLFVKGGQGRARGK 208
P SY H TL +G+DS+ LD + T L + + E+ +SG+ L ARGK
Sbjct: 485 PKSYSHFKETLLFGRDSVSLDEV-QTALNSKELNERKEKKSSASGEGL-------TARGK 330
Query: 209 GKAVDSG-KKKRSKSKDRKTAE-------CYSCKQIGHWKRDCPNRPGKSSNSN-----S 255
DS KK+ K +++K E CY CK+ GH ++ CP R ++N
Sbjct: 329 TFKKDSEFDKKKQKPENQKNGEGNIFKIRCYHCKKEGHTRKVCPERQKNGGSNNRKKDSG 150
Query: 256 SANVVQSDGSCSEEDLLCVSSVKCTDAWVLDSGCSYHMTPHREWFNSF 303
+A +VQ DG S E L+ VS W++DSGCS+HMTP++ WF F
Sbjct: 149 NAAIVQDDGYESAEALM-VSEKNPETKWIMDSGCSWHMTPNKSWFEQF 9
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 121 bits (303), Expect = 2e-27
Identities = 66/198 (33%), Positives = 111/198 (55%)
Frame = +1
Query: 875 FSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVC 934
F+PV R SIR +L + L QMDVK+ FL+G L E++Y+EQP+GF++ V
Sbjct: 3445 FAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVY 3624
Query: 935 KLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
+LK++LYGLKQ+PR Y+R ++ + GYR+ D ++V+ D + + +YVDD++
Sbjct: 3625 RLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQ-DAENLMIAQIYVDDIVF 3801
Query: 995 AANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLSRFD 1054
+ ++ EF+M +G LG+++ + + ++LSQ Y + ++ +F
Sbjct: 3802 GGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDS--IFLSQSRYAKNIVKKFG 3975
Query: 1055 MSKENPVSTPLANHFKLS 1072
M + TP H KLS
Sbjct: 3976 MENASHKRTPAPTHLKLS 4029
Score = 115 bits (289), Expect = 1e-25
Identities = 90/341 (26%), Positives = 161/341 (46%), Gaps = 23/341 (6%)
Frame = +1
Query: 219 RSKSKDRKTAECYSCKQIGHWKRDC------PNRPGKSSNSNSSANVVQSDGSCSEEDLL 272
+ K RK C+ C + GH K C P+ +SSNS V + S L+
Sbjct: 1477 QQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSNSRKKMMWVPKHKAVS---LV 1647
Query: 273 CVSSVKCT--DAWVLDSGCSYHMTPHREWFNSFKLGDFGYVYLGDDKPCIIKGMRQVKIA 330
+S++ + + W LDSGCS HMT +E+ + + YV GD I GM ++
Sbjct: 1648 VHTSLRASAKEDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKL--- 1818
Query: 331 LDDGRVRTLSQVRYVPEVTKNMISLGTLHENGYSFKSEENRDILRVSKGAMTVMRAKRTA 390
+ DG + +L++V V +T N+IS+ L + G++ ++ ++ K + +M+ R+
Sbjct: 1819 VHDG-LPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEV-LMKGSRSK 1992
Query: 391 GNIYKLLGGTVMGDVASVETDDDATKMWHMRLGHLSERGMMQLHKRNLLKGVRSCTI--- 447
N Y + + +D ++WH R GHL RGM ++ + ++G+ + I
Sbjct: 1993 DNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEG 2172
Query: 448 GLCKYCVLGKQCRVRFKTGQHKTKG-ILDYVHSDVWGPTKEPSVGGFRY-------FLHL 499
+C C +GKQ ++ + QH+T +L+ +H D+ GP + S+GG RY F
Sbjct: 2173 RICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRF 2352
Query: 500 LMIFLVRSGF----IF*NINLKSLPSSNCGRQRLRIRQGEK 536
+ +R +F ++L+ +C +R+R G +
Sbjct: 2353 TWVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGRE 2475
>CA937893 similar to GP|20805072|dbj retrovirus-related pol polyprotein from
transposon TNT 1-94-like, partial (7%)
Length = 412
Score = 120 bits (301), Expect = 4e-27
Identities = 57/104 (54%), Positives = 74/104 (70%)
Frame = -2
Query: 1 GAKFEVTRFYGTGNFGLWHRRVKDLLAQQSLQKALRDEKPTDIATVDWNEMKEKAASLIR 60
GAKFEV +F GTGNF LW +RVKDLLA Q L K LRD K + +DW E+ E+ A+ IR
Sbjct: 315 GAKFEVGKFDGTGNFRLWQKRVKDLLA*QGLLKVLRDSKSNNTEALDWEEL*ERTATTIR 136
Query: 61 LCVSDDLMNHILDLTTPKDVWDKLESLYMSKTLMNKLFAKQRLY 104
LC+ D+ + H+++L P +VW KLES YM K+L NKL+ Q+LY
Sbjct: 135 LCLVDEFLYHMMELAFPGEVWKKLESQYMLKSLTNKLYLMQKLY 4
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 119 bits (297), Expect = 1e-26
Identities = 66/198 (33%), Positives = 109/198 (54%)
Frame = +1
Query: 875 FSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVC 934
F+PV R SIR +L + L QMDVK+ FL+G L E+ Y+EQP+GF + V
Sbjct: 3448 FAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVY 3627
Query: 935 KLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLI 994
+LK++LYGLKQ+PR Y+R ++ + GYR+ D ++V+ D + + +YVDD++
Sbjct: 3628 RLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQ-DAENLMIAQIYVDDIVF 3804
Query: 995 AANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLSRFD 1054
+ ++ EF+M +G LG+++ + + ++LSQ Y + ++ +F
Sbjct: 3805 GGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDS--IFLSQSKYAKNIVKKFG 3978
Query: 1055 MSKENPVSTPLANHFKLS 1072
M + TP H KLS
Sbjct: 3979 MENASHKRTPAPTHLKLS 4032
Score = 114 bits (286), Expect = 2e-25
Identities = 88/338 (26%), Positives = 159/338 (47%), Gaps = 20/338 (5%)
Frame = +1
Query: 219 RSKSKDRKTAECYSCKQIGHWKRDCPN---RPGKSSNSNSSANVVQSDGSCSEEDLLCVS 275
+ K RK C+ C + GH K C + P + S+SS + L+ +
Sbjct: 1480 QQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSGRKMMWVPKHKIVSLVVHT 1659
Query: 276 SVKCT--DAWVLDSGCSYHMTPHREWFNSFKLGDFGYVYLGDDKPCIIKGMRQVKIALDD 333
S++ + + W LDSGCS HMT +E+ + + YV GD I GM ++ + D
Sbjct: 1660 SLRASAKEDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKL---VHD 1830
Query: 334 GRVRTLSQVRYVPEVTKNMISLGTLHENGYSFKSEENRDILRVSKGAMTVMRAKRTAGNI 393
G + +L++V V +T N+IS+ L + G++ ++ ++ K + +M+ R+ N
Sbjct: 1831 G-LPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEV-LMKGSRSKDNC 2004
Query: 394 YKLLGGTVMGDVASVETDDDATKMWHMRLGHLSERGMMQLHKRNLLKGVRSCTI---GLC 450
Y + + +D K+WH R GHL RGM ++ + ++G+ + I +C
Sbjct: 2005 YLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRIC 2184
Query: 451 KYCVLGKQCRVRFKTGQHKTKG-ILDYVHSDVWGPTKEPSVGGFRY-------FLHLLMI 502
C +GKQ ++ + QH+T +L+ +H D+ GP + S+GG RY F +
Sbjct: 2185 GECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWV 2364
Query: 503 FLVRSGF----IF*NINLKSLPSSNCGRQRLRIRQGEK 536
+R +F ++L+ +C +R+R G +
Sbjct: 2365 NFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGRE 2478
Score = 43.1 bits (100), Expect = 8e-04
Identities = 62/287 (21%), Positives = 103/287 (35%), Gaps = 49/287 (17%)
Frame = +1
Query: 33 KALRDEKPTDIATVDWNEM---KEKAASLIRLCVSDDLMNHILDLTTPKDVWDKLESLY- 88
K + KP + T + +E+ KA + + V ++ I T KD W+ L++ +
Sbjct: 172 KPTNELKPEEDWTKEEDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHE 351
Query: 89 -MSKTLMNKL-FAKQRLYSLKMQEGGDLQAHVYAFNNILADLTRLRVKVDDEDKAIILLR 146
SK M++L + +LKM+E + I T L ++ DE +LR
Sbjct: 352 GTSKVKMSRLQLLATKFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILR 531
Query: 147 SLPGSYDHLVTTLTYGKD--SIKLDSISSTLLQHAQRRQSVEEGGGSSGKCLFVKGGQGR 204
SLP +D VT + +D ++++D + +L E S FV +G
Sbjct: 532 SLPKRFDMKVTAIEEAQDICNMRVDELIGSLQTFELGLSDRTEK--KSKNLAFVSNDEGE 705
Query: 205 ARGKGKAVDSG-----------------------------------------KKKRSKSK 223
D G KK K
Sbjct: 706 EDEYDLDTDEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYQKKSDEKPS 885
Query: 224 DRKTAECYSCKQIGHWKRDCPNRPGKSSNSNSSANVVQSDGSCSEED 270
K +C+ C+ GH K +CP K +V +SD + SE++
Sbjct: 886 HSKGIQCHGCEGYGHIKAECPTHLKK---QRKGLSVCRSDDTESEQE 1017
>NP005470 reverse transcriptase
Length = 267
Score = 114 bits (284), Expect = 4e-25
Identities = 54/89 (60%), Positives = 65/89 (72%)
Frame = +1
Query: 904 KTTFLHGNLEEQIYMEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGY 963
+T FLHG LEE I M+QPEGF G R V +L+RSLYGLKQSPRQ Y FDS++ G+
Sbjct: 1 RTAFLHGRLEENILMKQPEGFEVQGKERYVSQLQRSLYGLKQSPRQWYMSFDSFITNQGF 180
Query: 964 RRCDYDCCVYVRSLDDGSFIFLLLYVDDM 992
+R YDCCVY ++DG I+LLLYVDDM
Sbjct: 181 KRSLYDCCVYHNKVEDGLMIYLLLYVDDM 267
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 94.0 bits (232), Expect = 4e-19
Identities = 49/94 (52%), Positives = 60/94 (63%)
Frame = -2
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
KG G+ FSPV + T++R LA+ A L Q+D+K FLHG+LEE IYMEQ
Sbjct: 301 KGYTQVYGIDYCDTFSPVAKLTTVRLFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQ 122
Query: 921 PEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRF 954
P GF G+ LVCKL RSLYGLKQSPR + +F
Sbjct: 121 PPGFVAQGEYGLVCKLHRSLYGLKQSPRAWFGKF 20
>CO983516
Length = 724
Score = 89.7 bits (221), Expect = 7e-18
Identities = 48/119 (40%), Positives = 73/119 (61%)
Frame = +2
Query: 875 FSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVC 934
F PV R SIR +L + L QMDVK+ FL+G L E++Y+EQP+GF + V
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 935 KLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDML 993
+LK++LYGLKQ+PR Y+R + + GYR+ D ++V+ D + + +YVDD++
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQ-DAENLMIAQIYVDDIV 718
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 88.2 bits (217), Expect = 2e-17
Identities = 59/154 (38%), Positives = 81/154 (52%), Gaps = 1/154 (0%)
Frame = +1
Query: 875 FSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQPEGFSETGD-GRLV 933
FSPV + + + ++ L +D K FLHG LEE++YMEQP GF G+ +V
Sbjct: 106 FSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFLHGYLEEEVYMEQPLGFVAQGESSNMV 285
Query: 934 CKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDML 993
C+L RS YGLKQSPR F I Y + D V+ G I+L++YVDD+
Sbjct: 286 CQLCRSFYGLKQSPRA--WPFLYCGAAIWYDSHEADHSVFYCHSPQGC-IYLIVYVDDIG 456
Query: 994 IAANHLHNVNELKTKLGKEFDMKDLGAAKKILGM 1027
I + H + LK L +F KDLG + LG+
Sbjct: 457 ITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>BE802806
Length = 285
Score = 87.8 bits (216), Expect = 3e-17
Identities = 42/82 (51%), Positives = 60/82 (72%)
Frame = -2
Query: 1015 MKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLSRFDMSKENPVSTPLANHFKLSLG 1074
MKDLG+A++ILG++IHRDR +L+LSQ +Y++ V+ RF M + PVSTPL +H KLS+
Sbjct: 284 MKDLGSARRILGIDIHRDRAKGELFLSQSNYLKKVVERFRMHQSKPVSTPLGHHTKLSVT 105
Query: 1075 QCSKTDSEIEGMSKIPYASAVG 1096
Q +T E M++ PYA+ VG
Sbjct: 104 QAPETAEERSKMNQTPYANGVG 39
>BM307983
Length = 406
Score = 82.4 bits (202), Expect = 1e-15
Identities = 36/68 (52%), Positives = 49/68 (71%)
Frame = +2
Query: 898 LEQMDVKTTFLHGNLEEQIYMEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSY 957
+ Q DVK FLHG+LEE++YME P G+ + G VC+LK++LYGLKQSPR + RF
Sbjct: 194 MHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGRFTQA 373
Query: 958 MLRIGYRR 965
ML +GY++
Sbjct: 374 MLSLGYKQ 397
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 82.0 bits (201), Expect = 2e-15
Identities = 41/99 (41%), Positives = 64/99 (64%)
Frame = -3
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
KG + + G+ ++PV R IR +LA V+ + L QMDVK+ FL+G ++E++Y+EQ
Sbjct: 302 KGYNQEEGIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQ 123
Query: 921 PEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYML 959
P GF V KL+++LYGLKQ+PR Y+R +++L
Sbjct: 122 PPGFEIPDKPTHVYKLQKALYGLKQAPRAWYERISNFLL 6
>TC232995
Length = 1009
Score = 78.6 bits (192), Expect = 2e-14
Identities = 42/151 (27%), Positives = 82/151 (53%)
Frame = +2
Query: 918 MEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSL 977
+EQP GF + V KL+++LYGLKQ+PR Y+R +++L + R D ++++
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKR- 178
Query: 978 DDGSFIFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKK 1037
+ + +YVDD++ + + E + EF+M +G K LG++I + +
Sbjct: 179 KHNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ--*G 352
Query: 1038 LWLSQKSYVEGVLSRFDMSKENPVSTPLANH 1068
++++Q Y + ++ RF M +STP++ +
Sbjct: 353 IFINQSKYCKELIKRFGMDSAKHMSTPMSTN 445
>BI317550 weakly similar to GP|9759590|dbj| polyprotein-like {Arabidopsis
thaliana}, partial (18%)
Length = 421
Score = 70.9 bits (172), Expect = 4e-12
Identities = 43/134 (32%), Positives = 74/134 (55%)
Frame = -2
Query: 938 RSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYVRSLDDGSFIFLLLYVDDMLIAAN 997
+SLYGLKQ+ R+ Y++ + +L+ GY + D ++ + + +F LL+YVDD+++A +
Sbjct: 420 KSLYGLKQASRKWYEKLTNLLLKEGYIQSISDYSLFTLTKGN-TFTALLVYVDDIILAGD 244
Query: 998 HLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLSRFDMSK 1057
+ + +K L F +K+LG K LG+E+ R + +SQ+ Y +L +
Sbjct: 243 SIDEFDRIKNVLDLAFKIKNLGKLKYFLGLEVAHSR--LGITISQRKYCLDLLKDSGLLG 70
Query: 1058 ENPVSTPLANHFKL 1071
P STPL KL
Sbjct: 69 CKPASTPLDTSIKL 28
>NP005250 reverse transcriptase
Length = 243
Score = 64.7 bits (156), Expect = 3e-10
Identities = 34/81 (41%), Positives = 46/81 (55%)
Frame = +1
Query: 907 FLHGNLEEQIYMEQPEGFSETGDGRLVCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRC 966
FL+G++EE IYM QPE F +VCKLK+S+Y LKQ RQ Y +F + G+
Sbjct: 4 FLNGDIEETIYMVQPENFVLGDSKSMVCKLKKSIYDLKQVSRQWYYKFHQVITSYGFEAN 183
Query: 967 DYDCCVYVRSLDDGSFIFLLL 987
D CVY ++FL+L
Sbjct: 184 VVDDCVY-HKFSGSKYVFLVL 243
>TC231744
Length = 794
Score = 63.5 bits (153), Expect = 6e-10
Identities = 54/239 (22%), Positives = 100/239 (41%), Gaps = 6/239 (2%)
Frame = +2
Query: 102 RLYSLKMQEGGDLQAHVYAFNNILADLTRLRVKVDDEDKAIILLRSLPGSYDHLVTTLTY 161
+L S+K + G+++ ++ +N+ + L L++++ ++ ++L SLP + +
Sbjct: 50 KLISMKYKGKGNIREYIMEISNLASKLKSLKLELGEDLFVHLVLISLPAHFGQFKVSYNT 229
Query: 162 GKDSIKLDSISSTLLQHAQR--RQSVEEGGGSSGKCLFVKGGQGRARGKGKAVDSGKKKR 219
KD L+ + S +Q +R R E K K D +K
Sbjct: 230 QKDKWSLNELISHCVQEEERL*RDRTESAHNKKRK---------------KTKDVAEKTS 364
Query: 220 SKSKDRKTAE--CYSCKQIGHWKRDCPNRPGKSSNSNSSANVVQSDGSCSEEDLLCVSSV 277
+ K +K E CY CK+ H K+ CP + +V CSE +L V
Sbjct: 365 *QKKQQKDEEFTCYFCKKSRHMKKKCPKYAAWRVKKDKFLTLV-----CSEVNLAFVPK- 526
Query: 278 KCTDAWVLDSGCSYH--MTPHREWFNSFKLGDFGYVYLGDDKPCIIKGMRQVKIALDDG 334
D W +DSG + H MT ++ D ++++GD K ++ + ++ L G
Sbjct: 527 ---DTWWVDSGATTHISMTMQGCLWSRLPSDDERFIFMGDGKKVAVEAIETFRLQLKIG 694
>BI321712
Length = 399
Score = 62.8 bits (151), Expect = 1e-09
Identities = 32/88 (36%), Positives = 50/88 (56%)
Frame = -3
Query: 985 LLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKS 1044
L LYVDD++ N+ E K + EF+M D+G LG+E+ ++ K ++++Q+
Sbjct: 385 LCLYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQE--DKGIFITQEG 212
Query: 1045 YVEGVLSRFDMSKENPVSTPLANHFKLS 1072
Y + VL +F M NPV TP+ KLS
Sbjct: 211 YAKEVLKKFKMDDANPVGTPMECGSKLS 128
>TC232593 weakly similar to UP|Q9XG91 (Q9XG91) Tpv2-1c protein (Fragment),
partial (16%)
Length = 562
Score = 60.5 bits (145), Expect = 5e-09
Identities = 36/119 (30%), Positives = 62/119 (51%), Gaps = 3/119 (2%)
Frame = +1
Query: 982 FIFLLLYVDDMLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLS 1041
F+ + LYVDD+L+ + V E K ++ + F+M +LG LG+EI + + K+ +
Sbjct: 190 FLIVSLYVDDLLVTRDDARLVEEFKQEMMQAFEMTNLGLMTYFLGIEIKQSQ--NKVLIC 363
Query: 1042 QKSYVEGVLSRFDMSKENPVSTPLANHFKLSLGQCSKTDSEIEGMSKIP---YASAVGC 1097
Q+ Y + +L +F M + VSTP+ K ++++G KI Y S +GC
Sbjct: 364 QRKYAKEILKKFQMEECKSVSTPMNQKEKF---------NKVDGADKIDEGYYRSLIGC 513
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 58.2 bits (139), Expect = 2e-08
Identities = 30/61 (49%), Positives = 37/61 (60%)
Frame = +3
Query: 861 KGIHSKRGLIMMRFFSPVVRHTSIRAVLALVASRDMHLEQMDVKTTFLHGNLEEQIYMEQ 920
KG G+ FSPV T++R LA+ A R L Q+D+K FLHG+LEE IYMEQ
Sbjct: 222 KGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLHGDLEEDIYMEQ 401
Query: 921 P 921
P
Sbjct: 402 P 404
>BU083646 weakly similar to GP|6642775|gb| gag-pol polyprotein {Vitis
vinifera}, partial (19%)
Length = 428
Score = 55.8 bits (133), Expect = 1e-07
Identities = 34/119 (28%), Positives = 59/119 (49%), Gaps = 9/119 (7%)
Frame = +1
Query: 273 CVSSVKCTDAWVLDSGCSYHMTPHREWFNSFKLGDFGYVYLGDDKPCIIKGMRQVKIALD 332
C ++ T +W++DSGC+ HMT RE F F V + ++ +KG V I
Sbjct: 43 CFAASSST*SWLIDSGCTNHMTYDRELFTELDEVVFSKVKIRNEAYIDVKGKETVAI*GH 222
Query: 333 DGRVRTLSQVRYVPEVTKNMISLGTLHENGYS---------FKSEENRDILRVSKGAMT 382
G ++ +S V YV E+++N++S+ L + GY K E+R++ + M+
Sbjct: 223 TG-LKLISNVLYVSEISQNLLSVPQLLKKGYKVLFEDKNCMIKDSESREVFNIQMKGMS 396
>BM143109
Length = 415
Score = 55.1 bits (131), Expect = 2e-07
Identities = 33/127 (25%), Positives = 67/127 (51%), Gaps = 1/127 (0%)
Frame = +1
Query: 933 VCKLKRSLYGLKQSPRQ*YKRFDSYMLRIGYRRCDYDCCVYV-RSLDDGSFIFLLLYVDD 991
V KLK+ LYGLKQ+ R Y+ ++L G+ + D +++ + L+D + + +YVDD
Sbjct: 43 VFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLND--ILLVQIYVDD 216
Query: 992 MLIAANHLHNVNELKTKLGKEFDMKDLGAAKKILGMEIHRDRGTKKLWLSQKSYVEGVLS 1051
++ + + + + EF+M + LG++I + + +++SQ Y + ++
Sbjct: 217 IIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTK--NGIFISQSKYCKDLIH 390
Query: 1052 RFDMSKE 1058
RF M +
Sbjct: 391 RFGMEND 411
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.146 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,244,604
Number of Sequences: 63676
Number of extensions: 949701
Number of successful extensions: 7599
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 7355
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7555
length of query: 1309
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1200
effective length of database: 5,698,948
effective search space: 6838737600
effective search space used: 6838737600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0065.12


