

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0063.10
(565 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
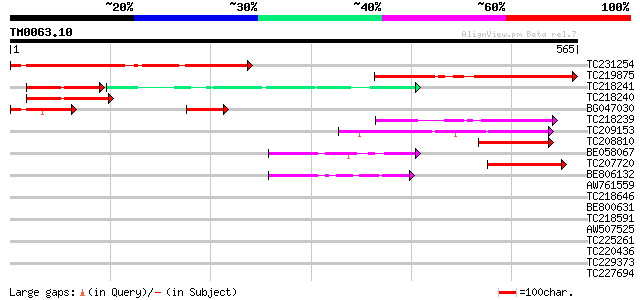
Score E
Sequences producing significant alignments: (bits) Value
TC231254 322 3e-88
TC219875 similar to UP|Q9VFF5 (Q9VFF5) CG17304-PA, partial (11%) 274 9e-74
TC218241 similar to UP|ROL_HUMAN (P14866) Heterogeneous nuclear ... 87 2e-36
TC218240 weakly similar to UP|Q79G09 (Q79G09) PE-PGRS FAMILY PRO... 91 2e-18
BG047030 89 4e-18
TC218239 61 2e-09
TC209153 similar to UP|YHGI_ECOLI (P46847) Protein yhgI, partial... 60 3e-09
TC208810 similar to PIR|D86350|D86350 F8K7.13 protein - Arabidop... 59 4e-09
BE058067 similar to PIR|D86350|D863 F8K7.13 protein - Arabidopsi... 59 4e-09
TC207720 similar to PIR|D86350|D86350 F8K7.13 protein - Arabidop... 59 8e-09
BE806132 similar to PIR|D86350|D863 F8K7.13 protein - Arabidopsi... 54 2e-07
AW761559 homologue to PIR|D86350|D863 F8K7.13 protein - Arabidop... 36 0.040
TC218646 weakly similar to UP|Q9FRR1 (Q9FRR1) F22O13.26 (Prenyla... 34 0.15
BE800631 weakly similar to GP|9294065|dbj| contains similarity t... 34 0.15
TC218591 weakly similar to UP|O65896 (O65896) Cytidine deaminase... 34 0.15
AW507525 similar to GP|6522580|emb| predicted protein {Arabidops... 33 0.26
TC225261 homologue to UP|RS14_LUPLU (O22584) 40S ribosomal prote... 33 0.34
TC220436 weakly similar to UP|Q50929 (Q50929) Opacity-related pr... 33 0.34
TC229373 similar to UP|Q6QAC9 (Q6QAC9) MYB transcription factor,... 33 0.44
TC227694 similar to UP|Q9SI52 (Q9SI52) Expressed protein (At2g03... 32 0.58
>TC231254
Length = 703
Score = 322 bits (825), Expect = 3e-88
Identities = 162/242 (66%), Positives = 185/242 (75%)
Frame = +2
Query: 1 MEVLKDPNSNPGRTDDSESELELYTIPSSSKWFAWDEIHETERTALKEFFDGTSISRTPK 60
MEV KDPNS DS+SELELYTIPSSS+WFAW+EIHETER A KE+FDG+SISR+PK
Sbjct: 38 MEVAKDPNSQA----DSDSELELYTIPSSSRWFAWEEIHETERAAFKEYFDGSSISRSPK 205
Query: 61 VYKEYRDFVINKYREDPSRKLTFTEVRKSLVGDVTLLRKVFLFLESCGLINYGVGEAEKE 120
+YKEYRDF+INKYRE+PSR+LTF+EVRKSLVGDVT L KVFLFLE LINYG
Sbjct: 206 IYKEYRDFIINKYREEPSRRLTFSEVRKSLVGDVTFLHKVFLFLEHWALINYGTA----- 370
Query: 121 EEREDEEEGEGERCKVRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSGGAGV 180
ED EE + CKVR EEGAP+GIRV ATPNSLKP+ PRN KS + GA +
Sbjct: 371 ---EDVEE---DHCKVRFEEGAPSGIRVAATPNSLKPMLLPRNGKSAAN-----ATGASL 517
Query: 181 KLPPLASFADVYGDLMSAKELNCGICGDKCGFEHYRSTKDNFTICMKCFKNGNYGEKRSV 240
KLPPLAS++DVYGDL+ KE NC +C +CG HYR T+DNF IC CFK+GNYGEKRS
Sbjct: 518 KLPPLASYSDVYGDLIRQKEGNCALCAHQCGSGHYRCTQDNFIICANCFKSGNYGEKRSA 697
Query: 241 ED 242
ED
Sbjct: 698 ED 703
>TC219875 similar to UP|Q9VFF5 (Q9VFF5) CG17304-PA, partial (11%)
Length = 804
Score = 274 bits (700), Expect = 9e-74
Identities = 150/202 (74%), Positives = 167/202 (82%)
Frame = +3
Query: 364 PSKRQRVAALSESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEED 423
PSKRQRVAALS+SSS LM QVGLIS VDPHITAAAADAA++ALCDE+LCPREIFDVEED
Sbjct: 3 PSKRQRVAALSDSSSLLMNQVGLISNVVDPHITAAAADAAVSALCDEDLCPREIFDVEED 182
Query: 424 YASSANSLISDSERAHEVEGLEMDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAAR 483
Y SA +L + EGLEM+RS+ +E IPL LR+RA ATALGAAAAR
Sbjct: 183 Y--SARALEGE-------EGLEMERSSLSE---------IPLTLRVRAATATALGAAAAR 308
Query: 484 AKLMADQEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSILTERI 543
AKL+ADQEDREIEHLVATIIEAQI K+ KVKHFD LELLM+KEHAE+E LKDSILTERI
Sbjct: 309 AKLLADQEDREIEHLVATIIEAQIEKMLRKVKHFDNLELLMEKEHAEMENLKDSILTERI 488
Query: 544 DVLQKTFRSGITRWKDYSYVKS 565
DVL++TFRSG+TRWKDYSY KS
Sbjct: 489 DVLRRTFRSGVTRWKDYSYAKS 554
>TC218241 similar to UP|ROL_HUMAN (P14866) Heterogeneous nuclear
ribonucleoprotein L (hnRNP L) (P/OKcl.14), partial (4%)
Length = 1206
Score = 86.7 bits (213), Expect(2) = 2e-36
Identities = 74/313 (23%), Positives = 126/313 (39%)
Frame = +2
Query: 97 LRKVFLFLESCGLINYGVGEAEKEEEREDEEEGEGERCKVRVEEGAPNGIRVVATPNSLK 156
+R+VF FLE+ GLINY + + + D++E
Sbjct: 452 VRRVFDFLETWGLINYHPSSSLTKPLKWDDKE---------------------------- 547
Query: 157 PLSAPRNAKSGGGGGGSVSGGAGVKLPPLASFADVYGDLMSAKELNCGICGDKCGFEHYR 216
KS + S A VK + K L C C C +
Sbjct: 548 -------TKSDSASNSTESSSAPVK--------------ENTKRL-CSGCKVVCTIACFA 661
Query: 217 STKDNFTICMKCFKNGNYGEKRSVEDFILNESNENSVKQSTVWTEGETLLLLESVLKHGD 276
K + T+C +C+ GNY + DF E +E + T W E ET LLE++ + D
Sbjct: 662 CDKYDLTLCARCYVRGNYRVGVNSSDFR*VEISEET---KTDWNEKETTNLLEAITHYSD 832
Query: 277 DWELVAQSVQTKTKLDCISKLIELPFGELMLGSAHRNDNINIANGIVNNAIQVQSSSSDH 336
DW+ V+Q V +T+ +C++ ++LPF ++ +N + + N V ++ ++
Sbjct: 833 DWKRVSQHVPGRTEKECVAHFLKLPFAN-QFQHYQQHPAVNGTDDVCNLLKMVTNADAES 1009
Query: 337 QETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSSSLMKQVGLISTSVDPHIT 396
+ + + PSKR R+ L+++S+ +M Q +S +
Sbjct: 1010ELDT------------------VASAEPSKRMRLTPLADASNPIMAQAAFLSALAGSEVA 1135
Query: 397 AAAADAAITALCD 409
AAA AA+T L +
Sbjct: 1136QAAAQAALTTLSE 1174
Score = 84.7 bits (208), Expect(2) = 2e-36
Identities = 38/78 (48%), Positives = 57/78 (72%)
Frame = +1
Query: 17 SESELELYTIPSSSKWFAWDEIHETERTALKEFFDGTSISRTPKVYKEYRDFVINKYRED 76
+ +E + +PS S+WF+WD I E E L EFF+ S S++P+VYK YR+ ++ +R +
Sbjct: 217 TSAEANVIVVPSYSRWFSWDSIDECEARHLPEFFE--SASKSPRVYKYYRNSIVKYFRYN 390
Query: 77 PSRKLTFTEVRKSLVGDV 94
P+RK+TFT+VRK+LVGDV
Sbjct: 391 PTRKITFTDVRKTLVGDV 444
>TC218240 weakly similar to UP|Q79G09 (Q79G09) PE-PGRS FAMILY PROTEIN,
partial (5%)
Length = 554
Score = 90.5 bits (223), Expect = 2e-18
Identities = 42/87 (48%), Positives = 63/87 (72%)
Frame = +2
Query: 17 SESELELYTIPSSSKWFAWDEIHETERTALKEFFDGTSISRTPKVYKEYRDFVINKYRED 76
+ +E + +PS S+WF+WD I E E L EFF+ S S++P+VYK YR+ ++ +R +
Sbjct: 299 ASAEANVIVVPSYSRWFSWDSIDECEVRHLPEFFE--SASKSPRVYKYYRNSIVKYFRYN 472
Query: 77 PSRKLTFTEVRKSLVGDVTLLRKVFLF 103
P+RK+TFT+VRK+LVGDV +R+VF F
Sbjct: 473 PTRKITFTDVRKTLVGDVGSIRRVFDF 553
>BG047030
Length = 422
Score = 89.4 bits (220), Expect = 4e-18
Identities = 49/89 (55%), Positives = 55/89 (61%), Gaps = 23/89 (25%)
Frame = +2
Query: 1 MEVLKDPNSNPGRTDDSESELELYTIPSSSK-----------------------WFAWDE 37
MEV KDPNS DS+SELELYTIPSSS WFAW+E
Sbjct: 32 MEVAKDPNSQA----DSDSELELYTIPSSSSK*ASIL*TLI*FEGI*AARFLAGWFAWEE 199
Query: 38 IHETERTALKEFFDGTSISRTPKVYKEYR 66
IHETER A KE+FDG+SISR+PK+YKE R
Sbjct: 200 IHETERAAFKEYFDGSSISRSPKIYKECR 286
Score = 62.0 bits (149), Expect = 7e-10
Identities = 26/42 (61%), Positives = 32/42 (75%)
Frame = +3
Query: 177 GAGVKLPPLASFADVYGDLMSAKELNCGICGDKCGFEHYRST 218
GA +KLPPLAS++DVYGDL+ KE NC +C +CG HYR T
Sbjct: 297 GASLKLPPLASYSDVYGDLIRQKEGNCALCAHQCGSGHYRCT 422
>TC218239
Length = 787
Score = 60.8 bits (146), Expect = 2e-09
Identities = 42/182 (23%), Positives = 84/182 (46%)
Frame = +1
Query: 365 SKRQRVAALSESSSSLMKQVGLISTSVDPHITAAAADAAITALCDENLCPREIFDVEEDY 424
SKR R+ L ++S+ +M Q +S + AAA AA+T L
Sbjct: 100 SKRMRLTPLXDASNPIMAQAAFLSALAGSEVAQAAAQAALTTL----------------- 228
Query: 425 ASSANSLISDSERAHEVEGLEMDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAARA 484
S+ +A ++ R+T + D G + + + + A
Sbjct: 229 --------SEVYKATKINYRAFPRNTLLQ--DAGITSN-------GGNTSDSFQGSRLHA 357
Query: 485 KLMADQEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSILTERID 544
+ ++E+ ++E ++ IIE Q+ +Q K+ HF++L+LLM+KE ++E++K+ +++
Sbjct: 358 NIQLEKEELDVEKAISEIIEVQMKNIQDKLVHFEDLDLLMEKEGQQMEQMKNMFFLDQLT 537
Query: 545 VL 546
+L
Sbjct: 538 LL 543
>TC209153 similar to UP|YHGI_ECOLI (P46847) Protein yhgI, partial (8%)
Length = 1330
Score = 60.1 bits (144), Expect = 3e-09
Identities = 58/233 (24%), Positives = 102/233 (42%), Gaps = 18/233 (7%)
Frame = +3
Query: 328 QVQSSSSDHQETSKTQNQTP-------------ELTSEKEPNGDAVNESPSKRQRVAALS 374
++ SSS QE + + P E + + E DA+ + SK Q + +
Sbjct: 60 EIPSSSDKAQEETLIEEPCPSVKDRHVSDSLPSETSKDAEMVSDAIPSTKSKPQNPESTN 239
Query: 375 ESSSSLMKQVGLISTS-VDPHITAAAADAAITALCDENLCPREIFDVEEDYASSANSLIS 433
+ SL ++ V + D+ + C DVE S +N ++S
Sbjct: 240 PAHESLETTDSVMDVDGVSNSLPLEKIDSQPLITSKSSQCNGTEKDVE--MMSPSNPVVS 413
Query: 434 DSERAHEVE---GLE-MDRSTQAEKDDRGPKDDIPLPLRLRAGIATALGAAAARAKLMAD 489
+S + G + D + E D K D RA ++T L AAAA+AKL+A+
Sbjct: 414 NSGAENGPNTGAGKDHADNGAKVEDDGTETKQDSSFEKVKRAAVST-LAAAAAKAKLLAN 590
Query: 490 QEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKDSILTER 542
QE+ +I L + +IE Q+ KL+ K+ F+++E ++ + +E + + ER
Sbjct: 591 QEEDQIRQLTSLLIEKQLHKLETKLAFFNDVENVVMRAREHVERSRHKLYHER 749
>TC208810 similar to PIR|D86350|D86350 F8K7.13 protein - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (11%)
Length = 836
Score = 59.3 bits (142), Expect = 4e-09
Identities = 29/75 (38%), Positives = 46/75 (60%)
Frame = +3
Query: 468 RLRAGIATALGAAAARAKLMADQEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKE 527
+++ L AAA +AKL +D E+REI+ L A I+ Q+ +L+ K+K F E+E L+ KE
Sbjct: 69 KVKDAAKAGLSAAAMKAKLFSDHEEREIQRLCANIVNHQLKRLELKLKQFAEIETLLMKE 248
Query: 528 HAEIEELKDSILTER 542
++E K I +R
Sbjct: 249 CEQLERTKQRIAADR 293
>BE058067 similar to PIR|D86350|D863 F8K7.13 protein - Arabidopsis thaliana,
partial (11%)
Length = 423
Score = 59.3 bits (142), Expect = 4e-09
Identities = 45/156 (28%), Positives = 76/156 (47%), Gaps = 5/156 (3%)
Frame = +3
Query: 259 WTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCISKLIELPFGELMLGSAHRNDNINI 318
WT+ ETLLLLE++ + ++W +A+ V TK+K CI + LP + L +NIN+
Sbjct: 27 WTDQETLLLLEAMEIYNENWNEIAEHVGTKSKAQCILHFLRLPMEDGKL------ENINV 188
Query: 319 AN-GIVNNAIQVQSSSSDH----QETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAAL 373
+ + +NAI S H +T+ T +QT D+ N P
Sbjct: 189 PSMSLSSNAINRDHSGRLHCYSNGDTAGTVHQT----------RDSDNRLP--------F 314
Query: 374 SESSSSLMKQVGLISTSVDPHITAAAADAAITALCD 409
+ S + +M V ++++V P + A+ A AA+ L +
Sbjct: 315 ANSGNPVMALVAFLASAVGPRVAASCAHAALAVLSE 422
>TC207720 similar to PIR|D86350|D86350 F8K7.13 protein - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (8%)
Length = 841
Score = 58.5 bits (140), Expect = 8e-09
Identities = 28/79 (35%), Positives = 50/79 (62%)
Frame = +2
Query: 477 LGAAAARAKLMADQEDREIEHLVATIIEAQIGKLQHKVKHFDELELLMKKEHAEIEELKD 536
L AAA +AKL AD E+REI+ L A I+ ++ +L+ K+K F E+E + +E ++E+++
Sbjct: 11 LSAAAMKAKLFADHEEREIQRLCANIVNNKLKRLELKLKQFAEIETQLMRECEQVEKVRQ 190
Query: 537 SILTERIDVLQKTFRSGIT 555
+ +ER ++ +G T
Sbjct: 191 RLASERSHIISTRLGNGGT 247
>BE806132 similar to PIR|D86350|D863 F8K7.13 protein - Arabidopsis thaliana,
partial (11%)
Length = 406
Score = 53.9 bits (128), Expect = 2e-07
Identities = 38/145 (26%), Positives = 70/145 (48%)
Frame = +3
Query: 259 WTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDCISKLIELPFGELMLGSAHRNDNINI 318
WT+ ETLLLLE+V + ++W +A+ V TK+K CI + LP + L +NIN+
Sbjct: 21 WTDQETLLLLEAVEVYNENWNEIAEHVGTKSKAQCILHFLRLPVEDGKL------ENINV 182
Query: 319 ANGIVNNAIQVQSSSSDHQETSKTQNQTPELTSEKEPNGDAVNESPSKRQRVAALSESSS 378
+++ + S+ + ++ + S + +G N S + + S +
Sbjct: 183 ------SSLSLLSNVKNQEDIGRL-----HCFSNGDSSGPVHNSQDSDGR--LPFTNSGN 323
Query: 379 SLMKQVGLISTSVDPHITAAAADAA 403
+M V ++++V P + A A AA
Sbjct: 324 PVMALVAFLASAVGPRVAATCAHAA 398
>AW761559 homologue to PIR|D86350|D863 F8K7.13 protein - Arabidopsis
thaliana, partial (7%)
Length = 429
Score = 36.2 bits (82), Expect = 0.040
Identities = 17/34 (50%), Positives = 23/34 (67%)
Frame = +1
Query: 477 LGAAAARAKLMADQEDREIEHLVATIIEAQIGKL 510
L AAA +AKL +D E+REI+ L A I+ Q+ L
Sbjct: 28 LSAAAMKAKLFSDHEEREIQRLCANIVNHQVSLL 129
>TC218646 weakly similar to UP|Q9FRR1 (Q9FRR1) F22O13.26 (Prenylated Rab
receptor 4) (At1g08770), partial (62%)
Length = 1127
Score = 34.3 bits (77), Expect = 0.15
Identities = 24/74 (32%), Positives = 32/74 (42%), Gaps = 9/74 (12%)
Frame = -2
Query: 115 GEAEKEEEREDEEEGEG---ERCKVRVEEGAPNGIRVVATPNS------LKPLSAPRNAK 165
G+ EE ED+EE +G E +V +G+ V L P +
Sbjct: 352 GDGVPEEAEEDDEERDGGVVEAEVAQVSSDPDHGVVVGVGAGEG*EV*KLAPWTGSHQGG 173
Query: 166 SGGGGGGSVSGGAG 179
SGGGGGG S G+G
Sbjct: 172 SGGGGGGGGSSGSG 131
>BE800631 weakly similar to GP|9294065|dbj| contains similarity to myb
proteins~gene_id:MRC8.8 {Arabidopsis thaliana}, partial
(8%)
Length = 413
Score = 34.3 bits (77), Expect = 0.15
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +2
Query: 259 WTEGETLLLLESVLKHGDDWELVAQSVQTKTKLDC 293
WTE E L L +V+KHG W +A+ V +T C
Sbjct: 29 WTEEEDLRLEAAVVKHGYCWSKIAEEVPPRTDSQC 133
>TC218591 weakly similar to UP|O65896 (O65896) Cytidine deaminase 1
(At2g19570) , partial (52%)
Length = 824
Score = 34.3 bits (77), Expect = 0.15
Identities = 26/66 (39%), Positives = 29/66 (43%)
Frame = -3
Query: 115 GEAEKEEEREDEEEGEGERCKVRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSV 174
G EK E E GER +VRV G + V LK L+A A GGG G V
Sbjct: 264 GFGEKVVGAEVVGEEVGERAEVRVFVGGDEDLDVGCVAEFLKELAA--MAAGGGGDGEGV 91
Query: 175 SGGAGV 180
G GV
Sbjct: 90 EAGLGV 73
>AW507525 similar to GP|6522580|emb| predicted protein {Arabidopsis
thaliana}, partial (9%)
Length = 443
Score = 33.5 bits (75), Expect = 0.26
Identities = 28/82 (34%), Positives = 34/82 (41%), Gaps = 3/82 (3%)
Frame = -1
Query: 134 CKVR---VEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSGGAGVKLPPLASFAD 190
C +R V G G+R A L+ P GGGGG SV G AG L A F
Sbjct: 296 CPIRRLLVLVGNVEGVRQQAAQRRQSNLAGPAAGLRGGGGGWSVGGAAGEGL-GAAEFRR 120
Query: 191 VYGDLMSAKELNCGICGDKCGF 212
V G + A E G + G+
Sbjct: 119 VGGGGIDAGETFAETGGTETGY 54
>TC225261 homologue to UP|RS14_LUPLU (O22584) 40S ribosomal protein S14,
complete
Length = 849
Score = 33.1 bits (74), Expect = 0.34
Identities = 16/29 (55%), Positives = 19/29 (65%)
Frame = +1
Query: 116 EAEKEEEREDEEEGEGERCKVRVEEGAPN 144
E E+E ERE E E E ER +V EEG+ N
Sbjct: 34 ERERERERERERERERERERVNAEEGSSN 120
>TC220436 weakly similar to UP|Q50929 (Q50929) Opacity-related protein
precursor (Fragment), partial (7%)
Length = 669
Score = 33.1 bits (74), Expect = 0.34
Identities = 23/63 (36%), Positives = 32/63 (50%)
Frame = -2
Query: 117 AEKEEEREDEEEGEGERCKVRVEEGAPNGIRVVATPNSLKPLSAPRNAKSGGGGGGSVSG 176
AE+E E+ EEE EGE + + EE ++ P ++K +S G GGS G
Sbjct: 374 AEEEREQVAEEESEGELSRSKTEE------KIFPAPEAMKREGEITKRESQLGLGGS-GG 216
Query: 177 GAG 179
GAG
Sbjct: 215 GAG 207
>TC229373 similar to UP|Q6QAC9 (Q6QAC9) MYB transcription factor, partial
(70%)
Length = 1250
Score = 32.7 bits (73), Expect = 0.44
Identities = 14/19 (73%), Positives = 15/19 (78%)
Frame = -1
Query: 118 EKEEEREDEEEGEGERCKV 136
E+E ERE E EGEGERC V
Sbjct: 128 EREREREREREGEGERCGV 72
>TC227694 similar to UP|Q9SI52 (Q9SI52) Expressed protein
(At2g03890/T18C20.9), partial (10%)
Length = 835
Score = 32.3 bits (72), Expect = 0.58
Identities = 30/85 (35%), Positives = 37/85 (43%), Gaps = 13/85 (15%)
Frame = +2
Query: 120 EEEREDEEEGEGERCKVRVEEGAPNGIRVVATPN-SLK-PLSAPRNAKSGGGGGGSVSGG 177
EEE E+EEE EGE + V A I V+ + SLK + +N GG V G
Sbjct: 116 EEEEEEEEEAEGESPQGFVTSAAEEKIPSVSELSVSLKNTMLDGKNQNQQKYSGGKVENG 295
Query: 178 -----------AGVKLPPLASFADV 191
A +LPP SF DV
Sbjct: 296 HFANTSSGHKSANEQLPPSISFVDV 370
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.130 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,038,809
Number of Sequences: 63676
Number of extensions: 289782
Number of successful extensions: 3455
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 2536
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3114
length of query: 565
length of database: 12,639,632
effective HSP length: 102
effective length of query: 463
effective length of database: 6,144,680
effective search space: 2844986840
effective search space used: 2844986840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0063.10


