

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.10
(306 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
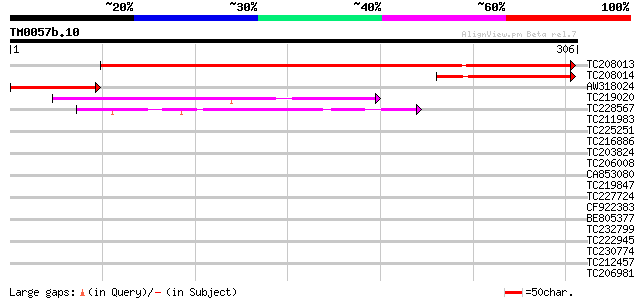
Score E
Sequences producing significant alignments: (bits) Value
TC208013 weakly similar to UP|Q8VYJ6 (Q8VYJ6) At2g30880/F7F1.9, ... 371 e-103
TC208014 similar to UP|Q8VYJ6 (Q8VYJ6) At2g30880/F7F1.9, partial... 111 4e-25
AW318024 similar to GP|17979458|gb| At2g30880/F7F1.9 {Arabidopsi... 87 9e-18
TC219020 similar to UP|SRB7_SCHPO (O94376) RNA polymerase II med... 45 5e-05
TC228567 41 8e-04
TC211983 39 0.003
TC225251 weakly similar to GB|AAO64828.1|29028898|BT005893 At3g5... 37 0.011
TC216886 weakly similar to UP|BIR3_HUMAN (Q13489) Baculoviral IA... 37 0.015
TC203824 similar to UP|Q9V3T8 (Q9V3T8) SR family splicing factor... 35 0.033
TC206008 similar to GB|AAO64828.1|29028898|BT005893 At3g56480 {A... 35 0.033
CA853080 weakly similar to GP|9759160|dbj| contains similarity t... 35 0.043
TC219847 35 0.056
TC227724 33 0.12
CF922383 33 0.12
BE805377 similar to GP|21648010|gb| peptidase M23/M37 family {C... 33 0.16
TC232799 weakly similar to UP|Q84TD8 (Q84TD8) At1g67170, partial... 33 0.16
TC222945 homologue to UP|Q8VAG3 (Q8VAG3) Wsv453 (WSSV513), parti... 32 0.47
TC230774 similar to UP|Q945L6 (Q945L6) AT3g11590/F24K9_26, parti... 32 0.47
TC212457 UP|Q39871 (Q39871) Maturation polypeptide, complete 31 0.80
TC206981 similar to UP|Q9LQB4 (Q9LQB4) F4N2.3, partial (25%) 30 1.4
>TC208013 weakly similar to UP|Q8VYJ6 (Q8VYJ6) At2g30880/F7F1.9, partial
(45%)
Length = 1065
Score = 371 bits (952), Expect = e-103
Identities = 198/256 (77%), Positives = 220/256 (85%)
Frame = +2
Query: 50 STIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYE 109
STI+DIA+KLSETAEAAEAAASAA+T+DE R AC EIE L DSEKQ EL QKLKE
Sbjct: 2 STIKDIADKLSETAEAAEAAASAAYTMDEHRRIACAEIERLNNDSEKQQELFTQKLKESA 181
Query: 110 VKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAE 169
KI+ LSKE+EQL +QR+AA QEANMWRSEL KARE VILE A+ RAEEKVRVAEANAE
Sbjct: 182 EKIVGLSKEREQLIRQRDAAIQEANMWRSELAKAREHDVILEAAVVRAEEKVRVAEANAE 361
Query: 170 AKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEEN 229
+I+EAVQ ESAA+ EK+ELLAYV+ LKAQLQRQHID+T+VFEKTESCSDTKHVD TEEN
Sbjct: 362 TRIREAVQRESAALKEKEELLAYVNVLKAQLQRQHIDTTQVFEKTESCSDTKHVDPTEEN 541
Query: 230 VDKACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVASESE 289
VDKACLSVSR++P P AE VV M TDQV IQPVGDNEWSDIQATEAR+ADVREVA E++
Sbjct: 542 VDKACLSVSRAIPVP--AENVVHMATDQVNIQPVGDNEWSDIQATEARIADVREVAPETD 715
Query: 290 VSSLDIPVISQQGTNH 305
SSLDIPV+SQ GTNH
Sbjct: 716 GSSLDIPVVSQPGTNH 763
>TC208014 similar to UP|Q8VYJ6 (Q8VYJ6) At2g30880/F7F1.9, partial (12%)
Length = 524
Score = 111 bits (278), Expect = 4e-25
Identities = 57/75 (76%), Positives = 64/75 (85%)
Frame = +2
Query: 231 DKACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVASESEV 290
DKACLSVSR++P PA E VV M TDQV IQPVGDNEWSD QATEAR+ADVREVA E++
Sbjct: 2 DKACLSVSRAIPVPA--ENVVHMATDQVNIQPVGDNEWSDTQATEARIADVREVAPETDG 175
Query: 291 SSLDIPVISQQGTNH 305
SSLDIPV++Q GTNH
Sbjct: 176 SSLDIPVVNQPGTNH 220
>AW318024 similar to GP|17979458|gb| At2g30880/F7F1.9 {Arabidopsis thaliana},
partial (16%)
Length = 350
Score = 87.0 bits (214), Expect = 9e-18
Identities = 42/49 (85%), Positives = 44/49 (89%)
Frame = +3
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRD 49
MQISLRN LGMM N+TTDGPMDDLTIMKETL VKDEELQN +RDLR RD
Sbjct: 204 MQISLRNALGMMNNRTTDGPMDDLTIMKETLRVKDEELQNLSRDLRARD 350
>TC219020 similar to UP|SRB7_SCHPO (O94376) RNA polymerase II mediator
complex protein srb7, partial (14%)
Length = 1446
Score = 44.7 bits (104), Expect = 5e-05
Identities = 41/181 (22%), Positives = 79/181 (42%), Gaps = 4/181 (2%)
Frame = +2
Query: 24 LTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNA 83
L I+++ L + ++LQ+ DLR + +I EKL + E +
Sbjct: 296 LEILRDDLNLTQDKLQSTESDLREAELRESEIIEKLKASEENLVVRGRDIEETATRHSEL 475
Query: 84 CEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKE----KEQLSKQREAATQEANMWRSE 139
ESL +DSE++L+ + +K + ++ SL ++ +EQ++K E +T N +
Sbjct: 476 QLLHESLTRDSEQKLQEAIEKFNNKDSEVQSLLEKIKILEEQIAKAGEQSTSLKNEFEES 655
Query: 140 LGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQ 199
L K +L E ++ +AE+K ++ V +L +D+L+
Sbjct: 656 LSKLT--------SLESENEDLKRQILDAESKSSQSFSENELLVGTNIQLKTKIDELEES 811
Query: 200 L 200
L
Sbjct: 812 L 814
>TC228567
Length = 1531
Score = 40.8 bits (94), Expect = 8e-04
Identities = 52/194 (26%), Positives = 83/194 (41%), Gaps = 8/194 (4%)
Frame = +2
Query: 37 ELQNFARDLRTRDSTIRD---IAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLK-- 91
E Q + + ST+ A+K E A+ AE A A T A E+ ES K
Sbjct: 701 EAQEEVEQAKAKSSTLESSLLAAQKEIEAAKVAEMLARDAIT-------ALEKSESAKGN 859
Query: 92 ---KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGV 148
KDS + L+ L+EY K +EQ + + EAAT + + R ++ E
Sbjct: 860 KNDKDSSSMVTLT---LEEYHELSRRAYKAEEQANARIEAATSQIQIARESELRSLEKLE 1030
Query: 149 ILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDST 208
L L+ E +++A N+E +A +G+ A E + A Q+Q +T
Sbjct: 1031ELNEELSVRRESLKIATGNSE----KANEGKLAVEHELRTWRAE--------QKQQEKAT 1174
Query: 209 EVFEKTESCSDTKH 222
E+ E+T ++ H
Sbjct: 1175ELNEQTSDPTEPAH 1216
>TC211983
Length = 1042
Score = 38.9 bits (89), Expect = 0.003
Identities = 53/235 (22%), Positives = 104/235 (43%), Gaps = 17/235 (7%)
Frame = +3
Query: 28 KETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEI 87
+E + ++LQ + + + E+L E AE A + A T+ + A +EI
Sbjct: 3 REKIAELPKKLQQAVEEANQANLLAQAAREELRRIKEEAEQAKAGASTMQSKLLAAQKEI 182
Query: 88 ESLKKDSEKQLELSAQKLKEYEVKI-----------ISLSKEKE-QLSKQREAATQEANM 135
E+ + SE+ + + L+E E ++LS E+ LSKQ A Q+ANM
Sbjct: 183 EAARA-SERLAIAATKALQESESSSRNNNELDSSSWVTLSVEEYYNLSKQAHDAEQQANM 359
Query: 136 ----WRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLA 191
SE+ A+ES + L ++ + + + +A + + +QEL
Sbjct: 360 RVAAANSEIEIAKESELKTLEKLNDVNREMAARRESLKIAMDKAEKAREGKLGVEQEL-- 533
Query: 192 YVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLT-EENVDKACLSVSRSVPDPA 245
K +A+ ++Q ++ E ++ +K+ +++ E + + + +RS P PA
Sbjct: 534 --RKWRAEHEQQRRNAGESGQRGVVKQSSKNPEVSFERHKEANSIDQTRSGPIPA 692
>TC225251 weakly similar to GB|AAO64828.1|29028898|BT005893 At3g56480
{Arabidopsis thaliana;} , partial (48%)
Length = 1272
Score = 37.0 bits (84), Expect = 0.011
Identities = 37/143 (25%), Positives = 68/143 (46%), Gaps = 9/143 (6%)
Frame = +1
Query: 56 AEKLSETAEAAEAAASAAHTIDEQWRNACEEIESL-----KKDSEKQL---ELSAQKLKE 107
A KLSE A EAA+ H + ++ R+A E ++ K D E+ + E A +L +
Sbjct: 10 AAKLSEEARLREAASLEKHVLLKKLRDALESLKGRVAGRNKDDVEEAIAIVEALAVQLTQ 189
Query: 108 YEVKIISLSKEKEQLSKQREAATQEA-NMWRSELGKARESGVILEGALARAEEKVRVAEA 166
E ++I E ++L+ + A+++A + E AR A+ R EE ++ E
Sbjct: 190 REGELIQEKAEVKKLANFLKQASEDAKKLVDEERAFARAEIEDARAAVQRVEEALQEHER 369
Query: 167 NAEAKIKEAVQGESAAVSEKQEL 189
++A K+ ++ V E + +
Sbjct: 370 MSQASGKQDMEQLMKEVQEARRI 438
>TC216886 weakly similar to UP|BIR3_HUMAN (Q13489) Baculoviral IAP
repeat-containing protein 3 (Inhibitor of apoptosis
protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF
signaling complex protein 1) (IAP homolog C) (Apoptosis
inhibitor 2) (API2), partial (4%)
Length = 1137
Score = 36.6 bits (83), Expect = 0.015
Identities = 51/209 (24%), Positives = 87/209 (41%), Gaps = 14/209 (6%)
Frame = +1
Query: 27 MKETLLVKDEELQNFAR----DLRTRDSTIRDIAEKLSETAEAAEAAA-SAAHTIDEQWR 81
+++T + + E++N R L ++ +R + + +E EA+ SA+ ++
Sbjct: 1 LEDTTMKRLSEMENALRKASGQLDLGNAAVRRLETENAEMKAEMEASKLSASESV----- 165
Query: 82 NACEEIESL-KKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSEL 140
AC E+ KK +K L QK K + +S EKE++ K +E Q +
Sbjct: 166 TACLEVAKREKKCLKKLLAWEKQKAKLQQ----EISDEKEKILKTKEILVQIRQCQKEAE 333
Query: 141 GKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVS-----EKQELLAY--- 192
K +E E ALA EE+ EA AE+ K ++ + K +LL
Sbjct: 334 VKWKEELKAKEEALALVEEERHCKEA-AESNNKRKLEALRLKIEIDFQRHKDDLLRLEQE 510
Query: 193 VDKLKAQLQRQHIDSTEVFEKTESCSDTK 221
+ +LKA Q + + T C K
Sbjct: 511 LSRLKASAQSAELHNQSSTSPTSDCKGAK 597
>TC203824 similar to UP|Q9V3T8 (Q9V3T8) SR family splicing factor SC35
(CG5442-PB) (LD32469p), partial (6%)
Length = 1532
Score = 35.4 bits (80), Expect = 0.033
Identities = 39/172 (22%), Positives = 70/172 (40%), Gaps = 2/172 (1%)
Frame = +2
Query: 35 DEELQNFARDLRTRDSTIRDIAEKLS--ETAEAAEAAASAAHTIDEQWRNACEEIESLKK 92
+ EL + D +RD+ K+ ET E I+E+ + E E+ K
Sbjct: 629 ENELAGLKKVKAESDVRVRDLERKIGVLETKE-----------IEERNKRIRFEEETRDK 775
Query: 93 DSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEG 152
EK+ E+ + K E++ ++ K ++EL + + + LE
Sbjct: 776 IDEKEREIKGYRQKVEELEKVAAEK-------------------KTELDELVKQKLRLEE 898
Query: 153 ALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQH 204
AL +EEKV E++ KEA + E +S ++ + V+ + L H
Sbjct: 899 ALRESEEKVTGLESSILQLRKEAKEAERVIISLNEKAVETVESIDRGLNGVH 1054
>TC206008 similar to GB|AAO64828.1|29028898|BT005893 At3g56480 {Arabidopsis
thaliana;} , partial (18%)
Length = 795
Score = 35.4 bits (80), Expect = 0.033
Identities = 30/106 (28%), Positives = 50/106 (46%), Gaps = 8/106 (7%)
Frame = +2
Query: 19 GPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDE 78
GP I +E + D+ + RDL ++ A KLS A+ E A+ H + +
Sbjct: 269 GPSLKDVIEQEAFNLSDQHKRISVRDLASKFDKNLAAAAKLSNEAKLREVASLEGHVLLK 448
Query: 79 QWRNACEEIESL-----KKDSEK---QLELSAQKLKEYEVKIISLS 116
+ R+A E + K+D EK +E A KL + E ++I++S
Sbjct: 449 KLRDALESLRGRFAGINKEDVEKAISMVEALAVKLTQNEGELIAMS 586
>CA853080 weakly similar to GP|9759160|dbj| contains similarity to myosin
heavy chain~gene_id:MEE6.21 {Arabidopsis thaliana},
partial (10%)
Length = 583
Score = 35.0 bits (79), Expect = 0.043
Identities = 30/92 (32%), Positives = 45/92 (48%), Gaps = 2/92 (2%)
Frame = +1
Query: 36 EELQNFARDLRTRDSTIRDIAEKLSETA--EAAEAAASAAHTIDEQWRNACEEIESLKKD 93
E LQ R L T+ +T D EK + A EA+E A ++E+ EE+ES K
Sbjct: 181 ERLQEEFRRLSTQMTTTFDANEKATMKALTEASEVRAQKM-LLEEKVHKVKEEVESTKAG 357
Query: 94 SEKQLELSAQKLKEYEVKIISLSKEKEQLSKQ 125
E +L + ++ +V+I + E E SKQ
Sbjct: 358 YEVKLNQLSNQIDTMKVQIQQMLLEIEDKSKQ 453
>TC219847
Length = 994
Score = 34.7 bits (78), Expect = 0.056
Identities = 31/136 (22%), Positives = 59/136 (42%), Gaps = 1/136 (0%)
Frame = +2
Query: 52 IRDIAEKLSETAEA-AEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEV 110
I K+ + +A AE + A T++ +W+ E L+K +++ + + KE+E
Sbjct: 233 IDSTTSKIQQLQKAFAELESYRAVTLNLKWKELEEHFHGLEKSLKRRFDELEDQEKEFEN 412
Query: 111 KIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEA 170
K +++ ++ ++REAA + L + +E A+ A EK R + A
Sbjct: 413 K----TRKAREILEKREAAVFAKE--QDSLQRLQEKRDAASFAIVNAREKQRKISSRELA 574
Query: 171 KIKEAVQGESAAVSEK 186
+G V EK
Sbjct: 575 TFSNGGKGGMPGVEEK 622
>TC227724
Length = 799
Score = 33.5 bits (75), Expect = 0.12
Identities = 26/93 (27%), Positives = 48/93 (50%), Gaps = 4/93 (4%)
Frame = +2
Query: 36 EELQNFARDLRT---RDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWR-NACEEIESLK 91
+EL +D+ + R + +R I ++++E E + H + E + N+ E+ESL
Sbjct: 206 KELMAILKDVESAQLRVAWLRSIVDEITENIELIDE-----HCVAETAKANSDREVESLN 370
Query: 92 KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSK 124
K+ E LE+ AQK +E + +E+LS+
Sbjct: 371 KELESNLEILAQKEQEVTDIKTRIEAIRERLSE 469
>CF922383
Length = 691
Score = 33.5 bits (75), Expect = 0.12
Identities = 22/99 (22%), Positives = 41/99 (41%)
Frame = +1
Query: 13 TNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASA 72
T +++D P + +E + + ELQ L+T + + ++S T E E +
Sbjct: 394 TERSSDSPSREFLEAQEKIQELEVELQRLTESLKTSEHENDQLKGEISVTKEKLEESGKK 573
Query: 73 AHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVK 111
+D + E+I + +QL + L EVK
Sbjct: 574 YEELDLSHKKLQEQILEPENKYNQQLSTLEEALXSQEVK 690
>BE805377 similar to GP|21648010|gb| peptidase M23/M37 family {Chlorobium
tepidum TLS}, partial (4%)
Length = 323
Score = 33.1 bits (74), Expect = 0.16
Identities = 24/110 (21%), Positives = 50/110 (44%)
Frame = +1
Query: 15 KTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAH 74
KT + ++ + ++E+ KDE ++ A + + + D K ET + +
Sbjct: 4 KTEESKLETVQEVEESSDKKDEAVKEVAEENTETEKKVADADTKKEETEKVS-------- 159
Query: 75 TIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSK 124
I+E E IES K+ + E+S + + E+K + ++K ++ K
Sbjct: 160 -IEESPEEKKEVIESETKEIVAETEVSKKTNETEEIKSVETEEKKPEVEK 306
>TC232799 weakly similar to UP|Q84TD8 (Q84TD8) At1g67170, partial (53%)
Length = 867
Score = 33.1 bits (74), Expect = 0.16
Identities = 29/119 (24%), Positives = 54/119 (45%), Gaps = 17/119 (14%)
Frame = +3
Query: 109 EVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALA----RAEEKVRV- 163
E K+ S E ++L+ + + + R EL A+ +L ++ E+++RV
Sbjct: 315 EQKLASQHAEMQRLATENQRLAATHGVLRQELAAAQHELQMLHAHVSALKGEREQQIRVQ 494
Query: 164 ----AEANAEAKIKEAVQGE--------SAAVSEKQELLAYVDKLKAQLQRQHIDSTEV 210
++ AEAK E+V+ E V + EL++ L +LQR H D+ ++
Sbjct: 495 LEKISKMEAEAKGAESVKMELQQARGEAQNLVVSRDELVSKAQHLTQELQRVHADAVQI 671
>TC222945 homologue to UP|Q8VAG3 (Q8VAG3) Wsv453 (WSSV513), partial (7%)
Length = 831
Score = 31.6 bits (70), Expect = 0.47
Identities = 43/170 (25%), Positives = 72/170 (42%)
Frame = -1
Query: 28 KETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEI 87
+E ++ EEL+ T +S++ K E A+ E A A E+ ++A E
Sbjct: 822 QEEMIEAQEELEQAKAQSSTLESSLL-AKHKEIEAAKVVEMLARDAIITLEKSKSA--EG 652
Query: 88 ESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESG 147
KDS + L+ L+EY K +EQ + + EAA + + R ++ E
Sbjct: 651 NKNDKDSSSMVTLT---LEEYHELSRRAYKAEEQANMRIEAANSQIQIARESELRSLEKL 481
Query: 148 VILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLK 197
L L+ E + +A N K+A +G+ A E + A +K K
Sbjct: 480 KELNEELSVRRESLSIATEN----FKKANEGKLAVEHELRTWRAEQEKQK 343
>TC230774 similar to UP|Q945L6 (Q945L6) AT3g11590/F24K9_26, partial (43%)
Length = 1176
Score = 31.6 bits (70), Expect = 0.47
Identities = 40/183 (21%), Positives = 88/183 (47%)
Frame = +3
Query: 57 EKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLS 116
+K E EAA + + ++ + R +ESL K ++L + L + ++ S
Sbjct: 303 KKEQEIVEAAIESIAGELDVERKLRR---RLESLNKKLGRELADTKTSLLKVVKELESEK 473
Query: 117 KEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAV 176
+ +E + + + ++A+ +S++ K + + + + +E +++ + E + ++ +
Sbjct: 474 RAREIIEQVCDELARDADEDKSDIEKQKRVSTKVCEEVEKEKEIMQLTDRLREERAQKKL 653
Query: 177 QGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLS 236
S A + +E A VDKL+ QL+ + +V EK+ S + L++E + A LS
Sbjct: 654 ---SEAKYQLEEKNAAVDKLRNQLE-AFLGGKQVREKSRSST-----HLSDEEI-AAYLS 803
Query: 237 VSR 239
SR
Sbjct: 804 RSR 812
>TC212457 UP|Q39871 (Q39871) Maturation polypeptide, complete
Length = 1741
Score = 30.8 bits (68), Expect = 0.80
Identities = 41/170 (24%), Positives = 65/170 (38%), Gaps = 4/170 (2%)
Frame = +1
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLS----ETAEAAEAAASAAHTIDEQWRNACEEIES 89
K EE + A + T ++T E +TAE AEAA + A I ++ A E E+
Sbjct: 910 KKEETKEMASE--TAEATANKAGEMKEATKKKTAETAEAAKNKAGEIKDR---AAETAEA 1074
Query: 90 LKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVI 149
K + + E++ K E + + E +KQ+ A +E K SG
Sbjct: 1075 AKNKTAETAEVTKNKALEMKDAAKDRTAETTDAAKQKTAQAKENT-------KENVSGAG 1233
Query: 150 LEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQ 199
E K++ E K V+ E + E L D++ Q
Sbjct: 1234 ETARRKMEEPKLQGKEGYGGRGDKVVVKVEESRPGAIAETLKAADQIAGQ 1383
>TC206981 similar to UP|Q9LQB4 (Q9LQB4) F4N2.3, partial (25%)
Length = 1981
Score = 30.0 bits (66), Expect = 1.4
Identities = 47/203 (23%), Positives = 81/203 (39%), Gaps = 18/203 (8%)
Frame = +1
Query: 13 TNKTTDGPMDDLTIMKETL---LVKDEELQNFARDLRTRDSTIRDIA---EKLSETAEAA 66
T K+ DG + DL + V + L + +D S +DI EKL ET E
Sbjct: 1120 TLKSKDGNLFDLNYESAYVDEGQVDCDGLASNDQDCGLTASNEKDIINEREKLKETDEYK 1299
Query: 67 EAAASAAHTIDEQWRNACEEIESLKK--DSEKQLELSAQKLKEYEVKIISLSKEKEQLSK 124
+A + Q + EE++ L+K +EK+L ++ KE ++ K+ E++
Sbjct: 1300 QAMEEEWASRQRQLQIQAEEVQRLRKRRKAEKRLLDMQRRQKERIEEVRETQKKDEEVMN 1479
Query: 125 QREAATQEANMWRSEL--------GKARESGVILEGALARAEEKVRVAEANAEAKI--KE 174
+E E ++L R G+ + G+ +V A A K
Sbjct: 1480 LKEQLRVEIQKGLNQLEMRCHDMPSLLRGLGIQVGGSFIPLPNEVHAAYKRALLKFHPDR 1659
Query: 175 AVQGESAAVSEKQELLAYVDKLK 197
A + + A E +E + +LK
Sbjct: 1660 ASKTDVRAQVEAEEKFKLISRLK 1728
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.307 0.122 0.315
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,832,414
Number of Sequences: 63676
Number of extensions: 83991
Number of successful extensions: 430
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 420
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 425
length of query: 306
length of database: 12,639,632
effective HSP length: 97
effective length of query: 209
effective length of database: 6,463,060
effective search space: 1350779540
effective search space used: 1350779540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0057b.10


