

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
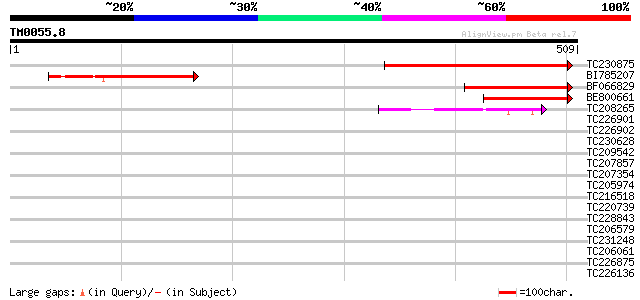
Query= TM0055.8
(509 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC230875 similar to GB|AAM91464.1|22137238|AY133634 AT5g14050/MU... 304 7e-83
BI785207 164 7e-41
BF066829 150 1e-36
BE800661 144 1e-34
TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%) 47 3e-05
TC226901 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia myo... 41 0.001
TC226902 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia myo... 40 0.003
TC230628 homologue to UP|Q9XIJ3 (Q9XIJ3) T10O24.21, partial (41%) 39 0.004
TC209542 38 0.012
TC207857 similar to UP|Q7ZY78 (Q7ZY78) Wdr4-prov protein, partia... 37 0.027
TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat prot... 35 0.080
TC205974 homologue to UP|Q94KS2 (Q94KS2) TGF-beta receptor-inter... 35 0.080
TC216518 weakly similar to UP|Q9LW87 (Q9LW87) Coatomer protein c... 35 0.080
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 34 0.18
TC228843 weakly similar to UP|BIN4_HUMAN (O15213) WD-repeat prot... 33 0.30
TC206579 weakly similar to UP|TAF5_YEAST (P38129) Transcription ... 33 0.30
TC231248 33 0.30
TC206061 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-... 33 0.40
TC226875 similar to UP|Q94JT6 (Q94JT6) At1g78070/F28K19_28, part... 33 0.40
TC226136 similar to UP|O48847 (O48847) Expressed protein (At2g32... 32 0.52
>TC230875 similar to GB|AAM91464.1|22137238|AY133634 AT5g14050/MUA22_5
{Arabidopsis thaliana;} , partial (39%)
Length = 1100
Score = 304 bits (778), Expect = 7e-83
Identities = 147/169 (86%), Positives = 159/169 (93%)
Frame = +3
Query: 337 SLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGG 396
SLEV E+S DSQMIAF GNEGYILLVSTKTKQLVG+LKM+GT RSLA++E+GQKLLS+GG
Sbjct: 375 SLEVLEVSPDSQMIAFTGNEGYILLVSTKTKQLVGTLKMNGTIRSLAYTENGQKLLSSGG 554
Query: 397 DGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKR 456
DGQVYHWDLRT TC+HKGVDEGCIN T LCTSPSGT FAAGSDSGIVN+YNRE+FLGGKR
Sbjct: 555 DGQVYHWDLRTMTCLHKGVDEGCINGTTLCTSPSGTYFAAGSDSGIVNIYNREEFLGGKR 734
Query: 457 KPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIPSYTVFSN 505
KPI+TIENL TKVDFMRFNHDSQILAICS MKKSSLKLIHIPSY+VFSN
Sbjct: 735 KPIRTIENLTTKVDFMRFNHDSQILAICSGMKKSSLKLIHIPSYSVFSN 881
>BI785207
Length = 421
Score = 164 bits (416), Expect = 7e-41
Identities = 95/143 (66%), Positives = 109/143 (75%), Gaps = 9/143 (6%)
Frame = +2
Query: 36 KRDRKREEHVVDMVEQVREMRKLESFLFGSLYSPLESGKEVDGEVEP-------SDLFFT 88
K+D +RE+ V EQV+EM+KLE+FLFGSLYSPL GK+ D EVEP SDLFFT
Sbjct: 2 KKDHEREQSTV---EQVKEMKKLENFLFGSLYSPLVFGKD-DDEVEPRKAAEKVSDLFFT 169
Query: 89 DRSADSVLSVC-DEDGEFSDGSGDGDDG-LGRKPAWVDEEEEKFTVNIAKVNRLRKLRKD 146
DRSADSVL+V +ED + S+GS D DD L RKP W+D+EEE TVNIAKVNRLRKLRK
Sbjct: 170 DRSADSVLTVYQEEDADLSEGSHDDDDRPLQRKPVWMDDEEENATVNIAKVNRLRKLRKV 349
Query: 147 EDESFISGSEYVARLRAHHVKLN 169
EDE ISG EYV+RLRA HVKLN
Sbjct: 350 EDEDLISGLEYVSRLRAQHVKLN 418
>BF066829
Length = 409
Score = 150 bits (379), Expect = 1e-36
Identities = 72/97 (74%), Positives = 81/97 (83%)
Frame = +2
Query: 409 TCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITK 468
TC+HKGV EGCIN T+LCTSPSGT FAA S SGIVN+YN + GGK KPI+TI+NL TK
Sbjct: 5 TCLHKGVHEGCINGTSLCTSPSGTHFAASSYSGIVNIYNITEIFGGKIKPIRTIDNLTTK 184
Query: 469 VDFMRFNHDSQILAICSSMKKSSLKLIHIPSYTVFSN 505
+DFM FNHDSQILAIC MKK+ LKLIHIPSY+V SN
Sbjct: 185 LDFMIFNHDSQILAICIGMKKTILKLIHIPSYSVISN 295
>BE800661
Length = 361
Score = 144 bits (362), Expect = 1e-34
Identities = 67/80 (83%), Positives = 78/80 (96%)
Frame = +2
Query: 426 CTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICS 485
CTSPSGT FAAGSDSGIVN+YN+++FLGGKRKPI+TIENL TKV+F++FNH+SQILAICS
Sbjct: 2 CTSPSGTHFAAGSDSGIVNIYNKKEFLGGKRKPIRTIENLTTKVNFIKFNHNSQILAICS 181
Query: 486 SMKKSSLKLIHIPSYTVFSN 505
SMKKSSLKLIHIPSY++FSN
Sbjct: 182 SMKKSSLKLIHIPSYSIFSN 241
>TC208265 similar to UP|Q6S7B0 (Q6S7B0) TAF5, partial (36%)
Length = 1048
Score = 46.6 bits (109), Expect = 3e-05
Identities = 41/163 (25%), Positives = 73/163 (44%), Gaps = 12/163 (7%)
Frame = +2
Query: 332 GREEKSLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMSGTTRSLAFSEDGQKL 391
G +K++ ++++ S + FVG+ G IL SLA S DG+ +
Sbjct: 293 GSSDKTVRLWDVQSGECVRVFVGHRGMIL--------------------SLAMSPDGRYM 412
Query: 392 LSAGGDGQVYHWDLRTRTCIHKGVDE-GCINSTALCTSPSGTLFAAGSDSGIVNVY---- 446
S DG + WDL + C+ + C+ S A S G++ A+GS V ++
Sbjct: 413 ASGDEDGTIMMWDLSSGRCLTPLIGHTSCVWSLAF--SSEGSVIASGSADCTVKLWDVNT 586
Query: 447 ----NREDFLGGKRKPIKTIENLITK---VDFMRFNHDSQILA 482
+R + GG +++++ L TK V +RF+ + + A
Sbjct: 587 STKVSRAEEKGGSANRLRSLKTLPTKSTPVYSLRFSRRNLLFA 715
>TC226901 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia
myotonica-containing WD repeat motif protein (DMR-N9
protein) (Protein 59) (Fragment), partial (13%)
Length = 1242
Score = 41.2 bits (95), Expect = 0.001
Identities = 24/79 (30%), Positives = 38/79 (47%), Gaps = 1/79 (1%)
Frame = +3
Query: 337 SLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLV-GSLKMSGTTRSLAFSEDGQKLLSAG 395
S++ S+D IA VG +GY+ + +QL+ G G A+S DG+ +L+ G
Sbjct: 129 SIDSISFSTDGTYIATVGRDGYLRVFDYLKEQLICGGKSYYGALLCCAWSMDGKYILTGG 308
Query: 396 GDGQVYHWDLRTRTCIHKG 414
D V W + R + G
Sbjct: 309 EDDLVQVWSMEDRKVVAWG 365
>TC226902 weakly similar to UP|DMWD_HUMAN (Q09019) Dystrophia
myotonica-containing WD repeat motif protein (DMR-N9
protein) (Protein 59) (Fragment), partial (14%)
Length = 1153
Score = 39.7 bits (91), Expect = 0.003
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 1/79 (1%)
Frame = +3
Query: 337 SLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLV-GSLKMSGTTRSLAFSEDGQKLLSAG 395
S+ S D +A VG +GY+ + +QL+ G G A+S DG+ +L+ G
Sbjct: 99 SINSISFSMDGAYLATVGRDGYLRVFDYSKEQLICGGKSYYGALLCCAWSMDGKYILTGG 278
Query: 396 GDGQVYHWDLRTRTCIHKG 414
D V W + R + G
Sbjct: 279 EDDLVQVWSMEDRKVVAWG 335
>TC230628 homologue to UP|Q9XIJ3 (Q9XIJ3) T10O24.21, partial (41%)
Length = 1119
Score = 39.3 bits (90), Expect = 0.004
Identities = 28/94 (29%), Positives = 46/94 (48%), Gaps = 11/94 (11%)
Frame = +2
Query: 337 SLEVFELSSDSQMIAFVGNEGYILLVSTKTKQLVGSLK------MSGTTRSLAFSEDGQK 390
S+ L ++ +A + IL+ ST+ K + K ++G + FS DGQ
Sbjct: 413 SMPSISLHPNANWLAAQSLDNQILIYSTREKFQLNKRKRFGGHIVAGYACQVNFSPDGQY 592
Query: 391 LLSAGGDGQVYHWDLRT----RTC-IHKGVDEGC 419
++S G+G+ + WD +T RT H+GV GC
Sbjct: 593 VMSGDGEGKCWFWDWKTCKVYRTLKCHEGVCIGC 694
Score = 34.7 bits (78), Expect = 0.10
Identities = 15/32 (46%), Positives = 18/32 (55%)
Frame = +2
Query: 376 SGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRT 407
S R + FS DG K LSAG D + +WD T
Sbjct: 20 SKAVRDICFSNDGTKFLSAGYDKNIKYWDTET 115
>TC209542
Length = 1445
Score = 37.7 bits (86), Expect = 0.012
Identities = 21/64 (32%), Positives = 34/64 (52%), Gaps = 1/64 (1%)
Frame = +2
Query: 380 RSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHK-GVDEGCINSTALCTSPSGTLFAAGS 438
R + ++D KL S GGD Q+++WD+ T I K +G +N + SG + +AG
Sbjct: 182 RDVHVTQDNSKLCSCGGDRQIFYWDVATGRVIRKFRGHDGEVNGVKFNENSSGVV-SAGY 358
Query: 439 DSGI 442
D +
Sbjct: 359 DQSL 370
Score = 32.7 bits (73), Expect = 0.40
Identities = 47/203 (23%), Positives = 88/203 (43%), Gaps = 3/203 (1%)
Frame = +2
Query: 296 ASFLPDGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSDSQMIAFVGN 355
A F DG+ V+ G+ + ++ +G K RE + + V + D+ + G
Sbjct: 62 ARFNGDGNYVLSCGKDRTIRLWNPHRGIHIKTYKSHAREVRDVHV---TQDNSKLCSCGG 232
Query: 356 EGYILLVSTKTKQLVGSLK-MSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTR-TCIHK 413
+ I T +++ + G + F+E+ ++SAG D + WD R+ T +
Sbjct: 233 DRQIFYWDVATGRVIRKFRGHDGEVNGVKFNENSSGVVSAGYDQSLRAWDCRSHSTEPIQ 412
Query: 414 GVDEGCINSTALCTSPSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMR 473
+D + ++C + T GS G V + D G+ +T +NL V+ +
Sbjct: 413 IIDTFADSVMSVCLTK--TEIIGGSVDGTVRTF---DIRIGR----ETSDNLGQPVNCVS 565
Query: 474 FNHDSQ-ILAICSSMKKSSLKLI 495
++D ILA C S+L+L+
Sbjct: 566 MSNDGNCILAGCLD---STLRLL 625
>TC207857 similar to UP|Q7ZY78 (Q7ZY78) Wdr4-prov protein, partial (8%)
Length = 887
Score = 36.6 bits (83), Expect = 0.027
Identities = 27/119 (22%), Positives = 53/119 (43%)
Frame = +3
Query: 380 RSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSD 439
R++ F G+ +SAG D + W ++ CI E + TA+ S GT
Sbjct: 165 RAIRFGAKGKLFVSAGDDKTLKVWSTQSWRCITTVSSEKRV--TAVAISDDGTFVCFADK 338
Query: 440 SGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIP 498
G+V V + + L +KP + + + + + F+ D + I S+ + +++ + P
Sbjct: 339 FGVVWVVDLDPPLHDDKKPAPLLSHYCSIITSLEFSPDGRY--ILSADRDFKIRVTNFP 509
>TC207354 weakly similar to UP|WDR5_HUMAN (P61964) WD-repeat protein 5,
partial (57%)
Length = 1124
Score = 35.0 bits (79), Expect = 0.080
Identities = 29/122 (23%), Positives = 56/122 (45%)
Frame = +1
Query: 384 FSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIV 443
F+ ++S D + WD++T C+H + + T++ + G L + S G
Sbjct: 415 FNPQSSYIVSGSFDETIKVWDVKTGKCVHT-IKGHTMPVTSVHYNRDGNLIISASHDGSC 591
Query: 444 NVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMKKSSLKLIHIPSYTVF 503
+++ E G K + IE+ V F +F+ + ++ I ++ +LKL + S VF
Sbjct: 592 KIWDTE--TGNLLKTL--IEDKAPAVSFAKFSPNGKL--ILAATLNDTLKLWNYGSGEVF 753
Query: 504 SN 505
N
Sbjct: 754 KN 759
>TC205974 homologue to UP|Q94KS2 (Q94KS2) TGF-beta receptor-interacting
protein 1, partial (57%)
Length = 783
Score = 35.0 bits (79), Expect = 0.080
Identities = 30/98 (30%), Positives = 41/98 (41%), Gaps = 4/98 (4%)
Frame = +3
Query: 350 IAFVGNEGYILLVSTKT-KQLVGSLKMSG---TTRSLAFSEDGQKLLSAGGDGQVYHWDL 405
I G + I + ++T K L S K SG T SLA S DG L+ D WD
Sbjct: 69 IISAGEDAVIRIWDSETGKLLKESDKESGHKKTVTSLAKSADGSHFLTGSLDKSARLWDT 248
Query: 406 RTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGIV 443
R+ T I V E +N+ + + G D+ V
Sbjct: 249 RSLTLIKTYVTERPVNAVTMSPLLDHVVIGGGQDASAV 362
>TC216518 weakly similar to UP|Q9LW87 (Q9LW87) Coatomer protein complex, beta
prime; beta'-COP protein, partial (29%)
Length = 1189
Score = 35.0 bits (79), Expect = 0.080
Identities = 50/245 (20%), Positives = 93/245 (37%), Gaps = 46/245 (18%)
Frame = +1
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V SV H +L + + K T+ +S+ + + P+R A F+ + ++ +
Sbjct: 103 VKSVDMHPTEPWILLGLYSGTISIWNYQTK--TEEKSLKISESPVRSAKFIARENWIVAA 276
Query: 309 GRRKFFYSFDLVKGTLEKIGPLVGREE--KSLEVFEL------SSDSQMIA--------- 351
K + ++ K +EKI ++ +SL V + +SD Q++
Sbjct: 277 TDDKNIHVYNYDK--MEKIVEFAEHKDYIRSLAVHPVLPYVISASDDQVLKLWNWRKGWS 450
Query: 352 ----FVGNEGYILLVSTKTKQ--------LVGSLKMSGTTRSLA---------------- 383
F G+ Y++ V+ K L G+LK+ S
Sbjct: 451 CYENFEGHSHYVMQVAFNPKDPSTFASASLDGTLKIWSLDSSAPNFTLEGHQKGVNCVDY 630
Query: 384 -FSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTSPSGTLFAAGSDSGI 442
+ D Q LLS D WD +R C+ + ++ N TA+C P + S+
Sbjct: 631 FITNDKQYLLSGSDDYTAKVWDYHSRNCV-QTLEGHENNVTAICAHPELPIIITASEDST 807
Query: 443 VNVYN 447
V +++
Sbjct: 808 VKIWD 822
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 33.9 bits (76), Expect = 0.18
Identities = 34/106 (32%), Positives = 45/106 (42%), Gaps = 7/106 (6%)
Frame = +1
Query: 373 LKMSGTTRS---LAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINST-ALCTS 428
L G +RS LAF DG S G D WDLRT I EG + + S
Sbjct: 427 LLQEGHSRSVYGLAFHNDGSLAASCGLDSLARVWDLRTGRSIL--ALEGHVKPVLGISFS 600
Query: 429 PSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTI---ENLITKVDF 471
P+G A G + +++ K+K TI NLI++V F
Sbjct: 601 PNGYHLATGGEDNTCRIWDLR-----KKKSFYTIPAHSNLISQVKF 723
Score = 28.5 bits (62), Expect = 7.5
Identities = 28/125 (22%), Positives = 50/125 (39%), Gaps = 1/125 (0%)
Frame = +1
Query: 241 IQDPSNGPVNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLP 300
+Q+ + V + FH + L + GLD R + D + I ++ P+ SF P
Sbjct: 430 LQEGHSRSVYGLAFHNDGSLAASCGLDSLARVW--DLRTGRSILALEGHVKPVLGISFSP 603
Query: 301 DGSQVILSGRRKFFYSFDLVKGTLEKIGPLVGREEKSLEVFELSSD-SQMIAFVGNEGYI 359
+G + G +DL R++KS S+ + F +EGY
Sbjct: 604 NGYHLATGGEDNTCRIWDL-------------RKKKSFYTIPAHSNLISQVKFEPHEGYF 744
Query: 360 LLVST 364
L+ ++
Sbjct: 745 LVTAS 759
>TC228843 weakly similar to UP|BIN4_HUMAN (O15213) WD-repeat protein BING4,
partial (5%)
Length = 1377
Score = 33.1 bits (74), Expect = 0.30
Identities = 12/30 (40%), Positives = 22/30 (73%)
Frame = +1
Query: 246 NGPVNSVQFHRNAQLLLTAGLDQKLRFFQI 275
+GPV+++ FH N L+ TAG D+K++ + +
Sbjct: 385 HGPVSALAFHSNGLLMATAGKDKKIKLWDL 474
Score = 30.4 bits (67), Expect = 2.0
Identities = 16/43 (37%), Positives = 22/43 (50%)
Frame = +1
Query: 364 TKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLR 406
T LV L G +LAF +G + +AG D ++ WDLR
Sbjct: 349 TSASPLVKMLCHHGPVSALAFHSNGLLMATAGKDKKIKLWDLR 477
>TC206579 weakly similar to UP|TAF5_YEAST (P38129) Transcription initiation
factor TFIID subunit 5 (TBP-associated factor 5)
(TBP-associated factor 90 kDa) (TAFII-90), partial (4%)
Length = 835
Score = 33.1 bits (74), Expect = 0.30
Identities = 45/205 (21%), Positives = 79/205 (37%), Gaps = 12/205 (5%)
Frame = +1
Query: 249 VNSVQFHRNAQLLLTAGLDQKLRFFQIDGKRNTKIQSIFLEDCPIRKASFLPDGSQVILS 308
V V H + L++ G + +R + + + + + D +R + + DGS V+ +
Sbjct: 19 VXXVVVHPDXXELISGGQNGNIRVWDLTAN-SCSCELVPEVDTAVRSLTVMWDGSLVVAA 195
Query: 309 GRRKFFYSFDLVKGT--------LEKIGPLVGREEKSLEVFELSSDSQMIAFVGNEGYIL 360
Y + L++GT L K+ G K L E + +A ++ +
Sbjct: 196 NNHGTCYVWRLLRGTQTMTNFEPLHKLQAHKGYILKCLLSPEFCEPHRYLATASSDHTVK 375
Query: 361 LVS----TKTKQLVGSLKMSGTTRSLAFSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVD 416
+ + T K L+G + FS DG L++A D W + T I V
Sbjct: 376 IWNVDGFTLEKTLIGHQRW---VWDCVFSVDGAYLITASSDTTARLWSMSTGEDIK--VY 540
Query: 417 EGCINSTALCTSPSGTLFAAGSDSG 441
+G +T C G A D+G
Sbjct: 541 QGHHKATICCALHDGAEPATS*DNG 615
>TC231248
Length = 849
Score = 33.1 bits (74), Expect = 0.30
Identities = 22/75 (29%), Positives = 31/75 (41%), Gaps = 4/75 (5%)
Frame = +1
Query: 377 GTTRSLAFSEDGQKLLSAGG-DGQVYHWDLRTRT-CIHK--GVDEGCINSTALCTSPSGT 432
G + FS DG L + G D + WD+R C++K E PSG
Sbjct: 160 GGITHVQFSRDGNYLYTGGRKDPYILCWDVRKSVDCVYKLYRSSENTNQRILFDIDPSGK 339
Query: 433 LFAAGSDSGIVNVYN 447
G G+V++YN
Sbjct: 340 YLGTGGQDGLVHIYN 384
>TC206061 weakly similar to UP|Q9FNN2 (Q9FNN2) WD-repeat protein-like,
partial (36%)
Length = 657
Score = 32.7 bits (73), Expect = 0.40
Identities = 21/90 (23%), Positives = 40/90 (44%), Gaps = 14/90 (15%)
Frame = +3
Query: 336 KSLEVFELSSDSQM-IAFVGNEGYILLVSTKTKQLVGSLKMSGT-------------TRS 381
++L++ E D + F N Y+ S ++ + M+G S
Sbjct: 183 RTLQILEAHDDEVWYVQFSHNGKYLASASNDRSAIIWEVDMNGELSVKHKLSGHQKPVSS 362
Query: 382 LAFSEDGQKLLSAGGDGQVYHWDLRTRTCI 411
+++S + Q+LL+ G + V WD+ T TC+
Sbjct: 363 VSWSPNDQELLTCGVEEAVRRWDVSTGTCL 452
>TC226875 similar to UP|Q94JT6 (Q94JT6) At1g78070/F28K19_28, partial (84%)
Length = 1542
Score = 32.7 bits (73), Expect = 0.40
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Frame = +1
Query: 343 LSSDSQMIAFVGNEGYILLVSTKTKQLVGSLKMS-GTTRSLAFSEDGQKLLSAGGDGQVY 401
+S D +++A +G+ L+ T ++ GSLK + S A+ DGQ L + D
Sbjct: 772 VSPDGKLLAVLGDSTECLIADANTGKITGSLKGHLDYSFSSAWHPDGQILATGNQDKTCR 951
Query: 402 HWDLR 406
WD+R
Sbjct: 952 LWDIR 966
>TC226136 similar to UP|O48847 (O48847) Expressed protein
(At2g32700/F24L7.16), partial (42%)
Length = 1463
Score = 32.3 bits (72), Expect = 0.52
Identities = 33/133 (24%), Positives = 55/133 (40%), Gaps = 1/133 (0%)
Frame = +3
Query: 370 VGSLKMSGTTRSLA-FSEDGQKLLSAGGDGQVYHWDLRTRTCIHKGVDEGCINSTALCTS 428
VGS++ S + FS DG+ L SAG D +V W++ T +E + T +
Sbjct: 210 VGSIRKSNSKVVCCHFSSDGKLLASAGHDKKVVLWNMETLQ-TESTPEEHSLIITDVRFR 386
Query: 429 PSGTLFAAGSDSGIVNVYNREDFLGGKRKPIKTIENLITKVDFMRFNHDSQILAICSSMK 488
P+ T A S V +++ D P+ T + V + F H + CS
Sbjct: 387 PNSTQLATSSFDTTVRLWDAAD----PTFPLHTYSGHTSHVVSLDF-HPKKTELFCSCDN 551
Query: 489 KSSLKLIHIPSYT 501
+ ++ I Y+
Sbjct: 552 NNEIRFWSISQYS 590
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.134 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,092,334
Number of Sequences: 63676
Number of extensions: 224178
Number of successful extensions: 1079
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 1046
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1071
length of query: 509
length of database: 12,639,632
effective HSP length: 101
effective length of query: 408
effective length of database: 6,208,356
effective search space: 2533009248
effective search space used: 2533009248
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0055.8


