

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0055.3
(355 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
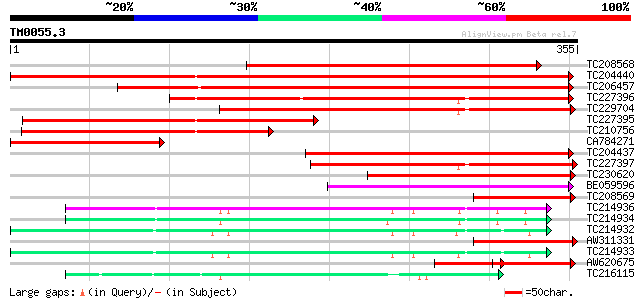
Score E
Sequences producing significant alignments: (bits) Value
TC208568 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {A... 309 1e-84
TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehy... 307 5e-84
TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannit... 277 5e-75
TC227396 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehy... 238 2e-63
TC229704 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehy... 228 4e-60
TC227395 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehy... 214 4e-56
TC210756 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehy... 179 2e-45
CA784271 164 4e-41
TC204437 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol de... 148 4e-36
TC227397 similar to UP|MTD1_STYHU (Q43137) Probable mannitol deh... 142 2e-34
TC230620 similar to UP|Q9ATW1 (Q9ATW1) Cinnamyl alcohol dehydrog... 121 4e-28
BE059596 110 1e-24
TC208569 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {A... 106 2e-23
TC214936 UP|O82478 (O82478) Alcohol dehydrogenase Adh-1 (Fragme... 85 4e-17
TC214934 homologue to UP|Q43016 (Q43016) Alcohol dehydrogenase-1... 83 2e-16
TC214932 homologue to UP|Q8LJR2 (Q8LJR2) Alcohol dehydrogenase 1... 83 2e-16
AW311331 80 1e-15
TC214933 UP|Q8LJR2 (Q8LJR2) Alcohol dehydrogenase 1 (Fragment), ... 80 2e-15
AW620675 similar to GP|13507210|gb| cinnamyl alcohol dehydrogena... 58 6e-12
TC216115 weakly similar to UP|ADHX_UROHA (P80467) Alcohol dehydr... 67 9e-12
>TC208568 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , partial (52%)
Length = 556
Score = 309 bits (791), Expect = 1e-84
Identities = 151/185 (81%), Positives = 171/185 (91%)
Frame = +1
Query: 149 LIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLNV 208
+IPKSYPLA+AAPLLCAGITVYSPM+RHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGL+V
Sbjct: 1 MIPKSYPLAAAAPLLCAGITVYSPMVRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLSV 180
Query: 209 TVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHSFDPYMALLKS 268
TVFSTSISKKEEALSLLGAD+FVVSS+QEEM ALAKSLDFI+DTASG HSFDPYM+LLK+
Sbjct: 181 TVFSTSISKKEEALSLLGADKFVVSSNQEEMTALAKSLDFIIDTASGDHSFDPYMSLLKT 360
Query: 269 FGVLALVGFPGEIKIHPGLLIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQ 328
+GV LVGFP ++K P L +GS+TV+GS GGTK+I++MI FC ANEIHPNIEV+PI+
Sbjct: 361 YGVFVLVGFPSQVKFIPASLNIGSKTVAGSVTGGTKDIQEMIGFCAANEIHPNIEVIPIE 540
Query: 329 YANEA 333
YANEA
Sbjct: 541 YANEA 555
>TC204440 similar to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (97%)
Length = 1481
Score = 307 bits (786), Expect = 5e-84
Identities = 152/354 (42%), Positives = 232/354 (64%), Gaps = 1/354 (0%)
Frame = +3
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
M S A +G AA D SG+LSPY ++ R G DDVY+K+ +CG+C++D+ +N LG
Sbjct: 60 MGSLEAERTTVGLAARDPSGILSPYTYNLRNTGPDDVYIKVHYCGICHSDLHQIKNDLGM 239
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 120
S YP VPGHE+ G V +VGS+V F VG+ VGVG V C++C+ C +E++C K ++
Sbjct: 240 SNYPMVPGHEVVGEVLEVGSDVSRFRVGELVGVGLLVGCCKNCQPCQQDIENYCSK-KIW 416
Query: 121 TFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQ 180
++N V DG T+GG+++ ++V +++ IP+ APLLCAG+TVYSP++ + +
Sbjct: 417 SYNDVYVDGKPTQGGFAETMIVEQKFVVKIPEGLAPEQVAPLLCAGVTVYSPLVHFGLKE 596
Query: 181 PGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMR 240
G G++GLGG+GHM VK KA G +VTV S+S KK+EAL LGAD+++VSSD M+
Sbjct: 597 SGLRGGILGLGGVGHMGVKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDATAMQ 776
Query: 241 ALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSG 299
A SLD+I+DT H +PY++LLK G L L+G ++ ++++G ++++GS
Sbjct: 777 EAADSLDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFVSPMVMLGRKSITGSF 956
Query: 300 VGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
+G KE +M+EF + IE+V + Y N+A +R+ K DV+YRFV+D++ S
Sbjct: 957 IGSMKETEEMLEFWKEKGLSSMIEMVNMDYINKAFERLEKNDVRYRFVVDVKGS 1118
>TC206457 weakly similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol
dehydrogenase (NAD-dependent mannitol dehydrogenase) ,
partial (75%)
Length = 1180
Score = 277 bits (709), Expect = 5e-75
Identities = 147/288 (51%), Positives = 195/288 (67%), Gaps = 2/288 (0%)
Frame = +3
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHEI G+VTKVG +V GF GD VGVG SC +CE C E++C K F +NGV
Sbjct: 27 GHEIIGVVTKVGRDVKGFKEGDRVGVGCLAASCLECEHCKTDQENYCEKLQ-FVYNGVFW 203
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKM-NQPGKSLG 186
DG+IT GGYS+ V RY IP++ + +AAPLLCAGITV++P+ H + PGK +G
Sbjct: 204 DGSITYGGYSQIFVADYRYVVHIPENLAMDAAAPLLCAGITVFNPLKDHDLVASPGKKIG 383
Query: 187 VIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSL 246
V+GLGGLGH+AVKFGKAFG +VTV STS SK+ EA LGAD F+VSS+ ++++A +S+
Sbjct: 384 VVGLGGLGHIAVKFGKAFGHHVTVISTSPSKEAEAKQRLGADDFIVSSNPKQLQAARRSI 563
Query: 247 DFIVDTASGPHSFDPYMALLKSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKE 305
DFI+DT S HS P + LLK G L LVG P + +++ LI G R+V G +GG KE
Sbjct: 564 DFILDTVSAEHSLLPILELLKVNGTLFLVGAPDKPLQLPAFPLIFGKRSVKGGIIGGIKE 743
Query: 306 IRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
++M+E C I +IE++ NEA++R+ K DV+YRFVIDI N+
Sbjct: 744 TQEMLEVCAKYNITSDIELITPDRINEAMERLAKNDVRYRFVIDIANA 887
>TC227396 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (70%)
Length = 927
Score = 238 bits (608), Expect = 2e-63
Identities = 123/256 (48%), Positives = 171/256 (66%), Gaps = 3/256 (1%)
Frame = +2
Query: 101 RDCEFCDDGLEHHCVKGSVFTFNGVDHDGTITKGGYSKFIVVHERYCFLIPKSYPLASAA 160
++CE C LE++C + VFT+N +DGT T+GGYS +VVH+RY P++ PL + A
Sbjct: 2 KECENCQQDLENYCPR-PVFTYNSPYYDGTRTQGGYSNIVVVHQRYVLRFPENLPLDAGA 178
Query: 161 PLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEE 220
PLLCAGITVYSPM + M +P ++GV GL GLGH+A+K KAFGL VTV S+S +K+ E
Sbjct: 179 PLLCAGITVYSPMKYYGMTEPA-NIGVEGLVGLGHVAIKLAKAFGLKVTVISSSPNKQAE 355
Query: 221 ALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPG- 279
A+ LGAD F+VSSD +M+ ++D+I+DT S HS P + LLK G L VG P
Sbjct: 356 AIDRLGADSFLVSSDPAKMKVALGTMDYIIDTISAVHSLIPLLGLLKLNGKLVTVGLPNK 535
Query: 280 --EIKIHPGLLIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRV 337
E+ I P L+ G + + GS GG KE ++M++FC + I +IE++ + N A++R+
Sbjct: 536 PLELPIFP--LVAGRKLIGGSNFGGIKETQEMLDFCAKHNITADIELIKMDQINTAMERL 709
Query: 338 IKKDVKYRFVIDIENS 353
K DVKYRFVID+ NS
Sbjct: 710 SKADVKYRFVIDVANS 757
>TC229704 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (63%)
Length = 816
Score = 228 bits (580), Expect = 4e-60
Identities = 117/226 (51%), Positives = 158/226 (69%), Gaps = 3/226 (1%)
Frame = +2
Query: 132 TKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLG 191
T GGYS +V E + IP PL +AAPLLCAGITVYSP+ + +++PG +GV+GLG
Sbjct: 2 TYGGYSDSMVADEHFVVRIPDRLPLDAAAPLLCAGITVYSPLRYYGLDKPGLHVGVVGLG 181
Query: 192 GLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVD 251
GLGHMAVKF KAFG VTV STS +KKEEA+ LGAD F++S DQ++M+A +LD I+D
Sbjct: 182 GLGHMAVKFAKAFGAKVTVISTSPNKKEEAIQNLGADSFLISRDQDQMQAAMGTLDGIID 361
Query: 252 TASGPHSFDPYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGSGVGGTKEIRD 308
T S H P + LLKS G L +VG P E+ + P L+ G + V+G+ +GG E ++
Sbjct: 362 TVSAVHPLLPLIGLLKSHGKLVMVGAPEKPLELPVFP--LLAGRKIVAGTLIGGLMETQE 535
Query: 309 MIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
MI+F + + P+IEV+P+ Y N A++R++K DVKYRFVIDI N+L
Sbjct: 536 MIDFAAKHNVKPDIEVIPMDYVNTAMERLLKADVKYRFVIDIGNTL 673
>TC227395 similar to UP|MTD_MEDSA (O82515) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (52%)
Length = 1002
Score = 214 bits (546), Expect = 4e-56
Identities = 102/185 (55%), Positives = 131/185 (70%)
Frame = +1
Query: 9 ECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVPG 68
+ GWAA+DTSG L+P+ FSRR G DDV +KI CGVC++D+ +N G + YP VPG
Sbjct: 67 KAFGWAASDTSGTLAPFHFSRRENGVDDVTLKILFCGVCHSDLHTLKNDWGFTTYPVVPG 246
Query: 69 HEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHD 128
HEI G+VT+VG+NV F VGD VGVG V SC++CE C LE++C + VFT+N +D
Sbjct: 247 HEIVGVVTEVGNNVKNFKVGDKVGVGVIVESCKECENCQQDLENYCPR-PVFTYNSPYYD 423
Query: 129 GTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVI 188
GT T+GGYS +VVH+RY P++ PL + APLLCAGIT YSPM + M +PGK LGV
Sbjct: 424 GTRTQGGYSNIMVVHQRYVLRFPENLPLDAGAPLLCAGITXYSPMKYYGMTEPGKHLGVA 603
Query: 189 GLGGL 193
LGG+
Sbjct: 604 RLGGV 618
>TC210756 similar to UP|MTD_FRAAN (Q9ZRF1) Probable mannitol dehydrogenase
(NAD-dependent mannitol dehydrogenase) , partial (45%)
Length = 549
Score = 179 bits (453), Expect = 2e-45
Identities = 82/158 (51%), Positives = 109/158 (68%)
Frame = +3
Query: 8 EECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGNSVYPCVP 67
++ GWAA D+SG+LSP+ FSRR G+ D+ K+ +CG+C++D+ +N GN+ YP VP
Sbjct: 78 KKVFGWAARDSSGLLSPFNFSRRETGEKDLVFKVQYCGICHSDLHMLKNEWGNTTYPLVP 257
Query: 68 GHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDH 127
GHEIAG+VT+VGS V F VGD VGVG + SCR CE CD+ LE++C K + T+
Sbjct: 258 GHEIAGVVTEVGSKVQKFKVGDRVGVGCMIGSCRSCESCDENLENYCPK-MILTYGVKYF 434
Query: 128 DGTITKGGYSKFIVVHERYCFLIPKSYPLASAAPLLCA 165
DGTIT GGYS +V E + IP + PL +AAPLLCA
Sbjct: 435 DGTITHGGYSDLMVADEHFVVRIPDNLPLDAAAPLLCA 548
>CA784271
Length = 436
Score = 164 bits (416), Expect = 4e-41
Identities = 75/97 (77%), Positives = 83/97 (85%)
Frame = +3
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
MSS+G E+CLGWAA D SGVLSPYKFSRR G++DV +KI+HCGVC+ADV WTRN G+
Sbjct: 144 MSSKGVGEDCLGWAARDASGVLSPYKFSRRTPGNEDVLIKITHCGVCFADVVWTRNKHGD 323
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYV 97
S YP VPGHEIAGIVTKVGSNVH F VGDHVGVGTYV
Sbjct: 324 SKYPVVPGHEIAGIVTKVGSNVHRFKVGDHVGVGTYV 434
>TC204437 homologue to UP|CADH_MEDSA (P31656) Cinnamyl-alcohol dehydrogenase
(CAD) , partial (49%)
Length = 844
Score = 148 bits (373), Expect = 4e-36
Identities = 76/169 (44%), Positives = 113/169 (65%), Gaps = 1/169 (0%)
Frame = +3
Query: 186 GVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKS 245
G++GLGG+GHM VK KA G +VTV S+S KK+EAL LGAD+++VSSD M+ A S
Sbjct: 21 GILGLGGVGHMGVKIAKALGHHVTVISSSDKKKQEALEHLGADQYLVSSDVTAMQEAADS 200
Query: 246 LDFIVDTASGPHSFDPYMALLKSFGVLALVG-FPGEIKIHPGLLIMGSRTVSGSGVGGTK 304
LD+I+DT H +PY++LLK G L L+G ++ ++++G R+++GS +G K
Sbjct: 201 LDYIIDTVPVGHPLEPYLSLLKLDGKLILMGVINTPLQFVSPMVMLGRRSITGSFIGSMK 380
Query: 305 EIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
E +M+EF + IEVV + Y N+A +R+ K DV+YRFV+D++ S
Sbjct: 381 ETEEMLEFWKEKGLSSMIEVVNMDYINKAFERLEKNDVRYRFVVDVKGS 527
>TC227397 similar to UP|MTD1_STYHU (Q43137) Probable mannitol dehydrogenase 1
(NAD-dependent mannitol dehydrogenase 1) , partial
(48%)
Length = 742
Score = 142 bits (358), Expect = 2e-34
Identities = 76/170 (44%), Positives = 107/170 (62%), Gaps = 3/170 (1%)
Frame = +2
Query: 189 GLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDF 248
GL GL H K KAFGL VTV S+S K+ EA+ LGAD F+VSSD +M+A ++D+
Sbjct: 2 GLXGLXHXXXKLAKAFGLKVTVISSSPXKQAEAIDRLGADFFLVSSDPAKMKAALGTMDY 181
Query: 249 IVDTASGPHSFDPYMALLKSFGVLALVGFPG---EIKIHPGLLIMGSRTVSGSGVGGTKE 305
I+DT S HS P + LLK G L VG P E+ I P L+ G + + GS GG KE
Sbjct: 182 IIDTISAVHSLIPLLGLLKLNGKLVTVGLPNKPLELPIFP--LVAGRKLIGGSNFGGLKE 355
Query: 306 IRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSLN 355
++M++FC + I +IE++ + N A++R+ + DVKYRFVID+ +S +
Sbjct: 356 TQEMLDFCGKHNITADIELIKMDQINTAMERLSRADVKYRFVIDVASSFS 505
>TC230620 similar to UP|Q9ATW1 (Q9ATW1) Cinnamyl alcohol dehydrogenase,
partial (37%)
Length = 690
Score = 121 bits (304), Expect = 4e-28
Identities = 60/131 (45%), Positives = 90/131 (67%), Gaps = 1/131 (0%)
Frame = +1
Query: 225 LGADRFVVSSDQEEMRALAKSLDFIVDTASGPHSFDPYMALLKSFGVLALVGFPGE-IKI 283
+GAD FVVS +Q++M+A+ ++D I+DT S H P + LLK G L +VG P + +++
Sbjct: 10 IGADSFVVSREQDQMQAVMGTMDGIIDTVSAVHPLVPLIGLLKPHGKLVMVGAPEKPLEL 189
Query: 284 HPGLLIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVK 343
L+MG + V GS +GG KE ++MI+F + + P+IEV+PI Y N A++R+ K D+K
Sbjct: 190 PVFSLLMGRKMVGGSSIGGMKETQEMIDFAAKHGVKPDIEVIPIDYVNTAIERLAKADLK 369
Query: 344 YRFVIDIENSL 354
YRFVIDI N+L
Sbjct: 370 YRFVIDIGNTL 402
>BE059596
Length = 466
Score = 110 bits (275), Expect = 1e-24
Identities = 64/155 (41%), Positives = 92/155 (59%), Gaps = 1/155 (0%)
Frame = +2
Query: 200 FGKAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASGPHSF 259
FGKAFG +VTV ST SK+ EA LGAD F++S + ++++A S+DFI+DT HS
Sbjct: 2 FGKAFGHHVTVISTCPSKEPEAKQRLGADHFILSCNPKQLQAARTSMDFILDTVVAEHSL 181
Query: 260 DPYMALLKSFGVLALVGFPGEIKIHPGL-LIMGSRTVSGSGVGGTKEIRDMIEFCVANEI 318
P + LLK G LVG P + P LI G R+V G +GG K+ +DM+E I
Sbjct: 182 LPILELLKVNGT*FLVGAPDKPLQSPAFPLIFGKRSVKGGIIGGIKKTQDMLEVRAKYNI 361
Query: 319 HPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENS 353
+IE+V +EA++R+ D + RF++ I N+
Sbjct: 362 RSDIELVTPHTIDEAMERLAHNDERCRFMLGIANA 466
>TC208569 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , partial (17%)
Length = 577
Score = 106 bits (264), Expect = 2e-23
Identities = 48/64 (75%), Positives = 59/64 (92%)
Frame = +3
Query: 291 GSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDI 350
GS+TV+GS GGTK+I++MI FC ANEIHPNIEV+PI+YANEAL+R+I +DVKYRFVID+
Sbjct: 96 GSKTVAGSVTGGTKDIQEMIGFCAANEIHPNIEVIPIEYANEALERLINRDVKYRFVIDV 275
Query: 351 ENSL 354
ENSL
Sbjct: 276 ENSL 287
>TC214936 UP|O82478 (O82478) Alcohol dehydrogenase Adh-1 (Fragment) ,
complete
Length = 1353
Score = 85.1 bits (209), Expect = 4e-17
Identities = 81/334 (24%), Positives = 139/334 (41%), Gaps = 30/334 (8%)
Frame = +3
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI + +C+ DV + ++P + GHE +GIV VG V GDH +
Sbjct: 138 EVRLKILYTSLCHTDVYFWDAKGQTPLFPRIFGHEASGIVESVGEGVTHLKPGDHA-LPV 314
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGVD-HDGT--ITKGG-----------YSKFIV 141
+ C DC C + C + T GV HDG +K G +S++ V
Sbjct: 315 FTGECGDCAHCKSEESNMCELLRINTDRGVMIHDGQSRFSKNGQPIHHFLGTSTFSEYTV 494
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH I + PL L C T + + +PG S+ + GLG + A +
Sbjct: 495 VHAGCVAKINPAAPLDKVCVLSCGICTGFGATVNVAKPKPGSSVAIFGLGAVALAAAEGA 674
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEE--MRALAKSLDFIVD-----TAS 254
+ G + + +S + E G + FV D ++ + +A+ + VD T S
Sbjct: 675 RVSGASRIIGVDLVSARFEEAKKFGVNEFVNPKDHDKPVQQVIAEMTNGGVDRAVECTGS 854
Query: 255 GPHSFDPYMALLKSFGVLALVGFPGE---IKIHPGLLIMGSRTVSGSGVGGTK---EIRD 308
+ + +G+ LVG P + K P + + RT+ G+ G K ++
Sbjct: 855 IQAMVSAFECVHDGWGLAVLVGVPSKDDAFKTAP-INFLNERTLKGTFYGNYKPRTDLPS 1031
Query: 309 MIEFCVANEIHPN---IEVVPIQYANEALDRVIK 339
++E ++ E+ + VP N+A D ++K
Sbjct: 1032VVEKYMSGELEVDKFITHTVPFSEINKAFDLMLK 1133
>TC214934 homologue to UP|Q43016 (Q43016) Alcohol dehydrogenase-1F ,
complete
Length = 1450
Score = 82.8 bits (203), Expect = 2e-16
Identities = 81/334 (24%), Positives = 134/334 (39%), Gaps = 30/334 (8%)
Frame = +3
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V +KI + +C+ DV + ++P + GHE GIV VG V GDH +
Sbjct: 201 EVRLKILYTSLCHTDVYFWEAKGQTPLFPRIFGHEAGGIVESVGEGVTHLKPGDHA-LPV 377
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGVD-HDGT-------------ITKGGYSKFIV 141
+ C +C C + C + T GV HD + +S++ V
Sbjct: 378 FTGECGECPHCKSEESNMCDLLRINTDRGVMIHDSQTRFSIKGQPIYHFVGTSTFSEYTV 557
Query: 142 VHERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFG 201
VH + + PL L C T + +PG S+ + GLG +G A +
Sbjct: 558 VHAGCVAKVNPAAPLDKICVLSCGICTGLGATINVAKPKPGSSVAIFGLGAVGLAAAEGA 737
Query: 202 KAFGLNVTVFSTSISKKEEALSLLGADRFVVSSD-----QEEMRALAK-SLDFIVD-TAS 254
+ G + + +S + E G + FV D QE + A+ +D V+ T S
Sbjct: 738 RISGASRIIGVDLVSSRFEEAKKFGVNEFVNPKDHDKPVQEVIAAMTNGGVDRAVECTGS 917
Query: 255 GPHSFDPYMALLKSFGVLALVGFPGE---IKIHPGLLIMGSRTVSGSGVGGTK---EIRD 308
+ + +GV LVG P + K HP + + RT+ G+ G K ++
Sbjct: 918 IQAMISAFECVHDGWGVAVLVGVPNKDDAFKTHP-VNFLNERTLKGTFYGNYKPRTDLPS 1094
Query: 309 MIEFCVANEIHPN---IEVVPIQYANEALDRVIK 339
++E + E+ VP N+A D ++K
Sbjct: 1095VVEKYMNGELELEKFITHTVPFSEINKAFDYMLK 1196
>TC214932 homologue to UP|Q8LJR2 (Q8LJR2) Alcohol dehydrogenase 1 (Fragment),
complete
Length = 1456
Score = 82.8 bits (203), Expect = 2e-16
Identities = 89/370 (24%), Positives = 141/370 (38%), Gaps = 31/370 (8%)
Frame = +3
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
MS+ G +C A + LS +V +KI +C DV W
Sbjct: 93 MSTAGQVIKCRAAVAWEAGKPLSIETIEVAPPQKGEVRLKILFNSLCRTDVYWWDAKGQT 272
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 120
++P + GHE +GIV VG V GDH + + C +C +C + C +
Sbjct: 273 PLFPRILGHEASGIVESVGKGVTHLKPGDH-ALPIFTGECGECTYCKSEESNLCELLRIN 449
Query: 121 TFNGV---DHDGTITKGG-----------YSKFIVVHERYCFLIPKSYPLASAAPLLCAG 166
T GV D +K G +S++ V+HE I + PL A + C
Sbjct: 450 TDRGVMLSDGKTRFSKNGQPIYHFVGTSTFSEYTVLHEGCVAKINPAAPLDKVAVVSCGF 629
Query: 167 ITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLG 226
T + + +P ++ V GLG +G A + + G + + ++ + E G
Sbjct: 630 CTGFGATVNVAKPKPNNTVAVFGLGAVGLAACEGARVSGASRIIGVDLLTNRFEQAKQFG 809
Query: 227 ADRFVVSSDQEE--MRALAKSLDFIVD-----TASGPHSFDPYMALLKSFGVLALVGFP- 278
FV D + +A+ + VD T S S + +G LV P
Sbjct: 810 VTDFVNPKDHNKPVQEVIAEMTNGGVDRAIECTGSIQASISAFECTHDGWGTAVLVSVPK 989
Query: 279 --GEIKIHPGLLIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEV-------VPIQY 329
E K HP + M RT+ G+ G + D I V ++ +E+ VP
Sbjct: 990 KDAEFKTHP-MKFMEGRTLKGTFYGHYRPRTD-IPGVVEKYLNKELELDKFITHSVPFSE 1163
Query: 330 ANEALDRVIK 339
N A D ++K
Sbjct: 1164INTAFDLMLK 1193
>AW311331
Length = 317
Score = 80.5 bits (197), Expect = 1e-15
Identities = 36/65 (55%), Positives = 48/65 (73%)
Frame = +1
Query: 291 GSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDI 350
G + V GS +GG KE ++MI+F + + P+IEV+PI Y N A+DR+ K DVKY FVIDI
Sbjct: 4 GRKMVGGSSIGGMKETQEMIDFAAKHGVKPDIEVIPIDYVNTAIDRLAKADVKYLFVIDI 183
Query: 351 ENSLN 355
EN+LN
Sbjct: 184 ENTLN 198
>TC214933 UP|Q8LJR2 (Q8LJR2) Alcohol dehydrogenase 1 (Fragment), complete
Length = 1477
Score = 79.7 bits (195), Expect = 2e-15
Identities = 88/370 (23%), Positives = 142/370 (37%), Gaps = 31/370 (8%)
Frame = +1
Query: 1 MSSEGASEECLGWAATDTSGVLSPYKFSRRAVGDDDVYVKISHCGVCYADVAWTRNTLGN 60
MS+ G +C A + LS +V ++I +C +DV W
Sbjct: 52 MSTAGQVIKCRAAVAWEAGKPLSIETIEVAPPQKGEVRLRILFNSLCRSDVYWWDAKDQT 231
Query: 61 SVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGTYVNSCRDCEFCDDGLEHHCVKGSVF 120
++P + GHE +GIV VG V GDH + + C +C +C + C +
Sbjct: 232 PLFPRILGHEASGIVESVGEGVTHLKPGDH-ALPIFTGECGECTYCKSEESNLCELLRIN 408
Query: 121 TFNGV---DHDGTITKGG-----------YSKFIVVHERYCFLIPKSYPLASAAPLLCAG 166
T GV D +K G +S++ V+HE I + PL A + C
Sbjct: 409 TDRGVMLSDGKTRFSKNGQPIYHFVGTSTFSEYTVLHEGCVAKINPNAPLDKVAIVSCGF 588
Query: 167 ITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFGKAFGLNVTVFSTSISKKEEALSLLG 226
T + + +P ++ V GLG +G A + + G + + + + E G
Sbjct: 589 CTGFGATVNVAKPKPNNTVAVFGLGAVGLAACEGARVSGASRIIGVDLLPNRFEQAKKFG 768
Query: 227 ADRFVVSSDQEE--MRALAKSLDFIVD-----TASGPHSFDPYMALLKSFGVLALVGFPG 279
FV D + +A+ + VD T S S + +G LVG P
Sbjct: 769 VTDFVNPKDHNKPVQEVIAEMTNGGVDRAIECTGSIQASISAFECTHDGWGTAVLVGVPK 948
Query: 280 ---EIKIHPGLLIMGSRTVSGSGVGGTKEIRDMIEFCVANEIHPNIEV-------VPIQY 329
E K +P + M RT+ G+ G + D I V ++ +E+ VP
Sbjct: 949 KDVEFKTNP-MKFMEGRTLKGTFYGHYRPRTD-IPGVVEKYLNKELELDKFITHSVPFSK 1122
Query: 330 ANEALDRVIK 339
N A D ++K
Sbjct: 1123INTAFDLMLK 1152
>AW620675 similar to GP|13507210|gb| cinnamyl alcohol dehydrogenase {Fragaria
x ananassa}, partial (24%)
Length = 406
Score = 57.8 bits (138), Expect(2) = 6e-12
Identities = 24/52 (46%), Positives = 38/52 (72%)
Frame = +2
Query: 303 TKEIRDMIEFCVANEIHPNIEVVPIQYANEALDRVIKKDVKYRFVIDIENSL 354
++E +++I+F + + P IEV+P+ Y N A++R++ DVKYRFVIDI N L
Sbjct: 119 SQETQEIIDFATEHNVRPQIEVIPMDYVNIAMERLLNADVKYRFVIDIGNIL 274
Score = 30.4 bits (67), Expect(2) = 6e-12
Identities = 16/45 (35%), Positives = 28/45 (61%), Gaps = 1/45 (2%)
Frame = +3
Query: 267 KSFGVLALVGFPGE-IKIHPGLLIMGSRTVSGSGVGGTKEIRDMI 310
KS G L +VG P + +++ LIMG +T++ S +GG K + ++
Sbjct: 9 KSHGKLVMVGAPEKPLELLLPSLIMGRKTIAASYIGGVKRHKKLL 143
>TC216115 weakly similar to UP|ADHX_UROHA (P80467) Alcohol dehydrogenase
class III (Glutathione-dependent formaldehyde
dehydrogenase) (FDH) , partial (42%)
Length = 1433
Score = 67.4 bits (163), Expect = 9e-12
Identities = 73/302 (24%), Positives = 120/302 (39%), Gaps = 28/302 (9%)
Frame = +1
Query: 36 DVYVKISHCGVCYADVAWTRNTLGNSVYPCVPGHEIAGIVTKVGSNVHGFSVGDHVGVGT 95
+V VK+ +C D++ T+ ++ +P GHE GI+ VG V GD V + T
Sbjct: 166 EVRVKMLCASICSTDISSTKG-FPHTNFPIALGHEGVGIIESVGDQVTNLKEGD-VVIPT 339
Query: 96 YVNSCRDCEFCDDGLEHHCVKGSVFTFNGVDHDGT-------------ITKGGYSKFIVV 142
Y+ C++CE C + C+ V + G+ D T + +S+++V
Sbjct: 340 YIGECQECENCVSEKTNLCMTYPV-RWTGLMPDNTSRMSIRGERIYHIFSCATWSEYMVS 516
Query: 143 HERYCFLIPKSYPLASAAPLLCAGITVYSPMMRHKMNQPGKSLGVIGLGGLGHMAVKFGK 202
Y + + A A+ + C T + + + G ++ V GLG +G AV K
Sbjct: 517 DANYVLKVDPTIDRAHASFISCGFSTGFGAAWKEAKVESGSTVAVFGLGAVGLGAVIGSK 696
Query: 203 AFGLNVTVFSTSISKKEEALSLLGADRFVVSSDQEEMRALAKSLDFIVDTASG----PHS 258
G + + + K G F+ D KS +V SG +S
Sbjct: 697 MQGASRIIGIDTNENKRAKGEAFGITDFINPGDSN------KSASELVKELSGGMGADYS 858
Query: 259 F----------DPYMALLKSFGVLALVGFPGEIKIHPGLL-IMGSRTVSGSGVGGTKEIR 307
F + A G ++G EI + GL I+ RT+ GS GG + I
Sbjct: 859 FECTGVSTLLSESLEATKIGTGKAIVIGVGIEITLPLGLFAILLGRTLKGSVFGGLRAIS 1038
Query: 308 DM 309
D+
Sbjct: 1039DL 1044
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,213,513
Number of Sequences: 63676
Number of extensions: 239181
Number of successful extensions: 1026
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 995
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1003
length of query: 355
length of database: 12,639,632
effective HSP length: 98
effective length of query: 257
effective length of database: 6,399,384
effective search space: 1644641688
effective search space used: 1644641688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0055.3


