

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
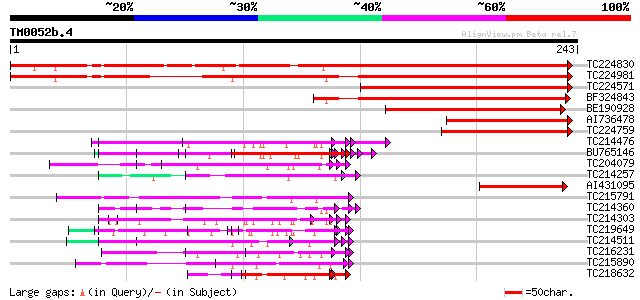
Query= TM0052b.4
(243 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224830 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-... 318 1e-87
TC224981 similar to UP|O50000 (O50000) Abscisic stress ripening ... 280 4e-76
TC224571 similar to UP|Q9ZRB6 (Q9ZRB6) Ci21A protein, complete 168 2e-42
BF324843 147 4e-36
BE190928 109 1e-24
AI736478 102 2e-22
TC224759 similar to UP|Q9SPF3 (Q9SPF3) Ripening induced protein ... 74 5e-14
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 73 1e-13
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 70 1e-12
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 69 1e-12
TC214257 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 68 3e-12
AI431095 weakly similar to PIR|T09789|T097 abscisic acid- and wa... 66 1e-11
TC215791 similar to GB|CAE76446.1|38567152|BX842633 related to p... 65 3e-11
TC214360 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding ... 65 4e-11
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 64 5e-11
TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich... 62 3e-10
TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protei... 62 3e-10
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 61 4e-10
TC215890 similar to PIR|F96770|F96770 protein RNA-binding protei... 61 4e-10
TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wal... 61 4e-10
>TC224830 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (61%)
Length = 1118
Score = 318 bits (815), Expect = 1e-87
Identities = 167/247 (67%), Positives = 188/247 (75%), Gaps = 6/247 (2%)
Frame = +3
Query: 1 MAEENRDRG-LFHHHKNED-RPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGY 58
MAEE + LFHHHK+ED +P +T+TGY+ TSYS +PS DYDSG+ K SYE+SGGGY
Sbjct: 171 MAEEKHHKHRLFHHHKDEDNKPVETDTGYDNTSYS--KPSD-DYDSGFNKPSYESSGGGY 341
Query: 59 ETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYS--TDEPSGGGYGTGGAGGGYSTGGGYGT 116
E GYNKTS YS+DEP SGGG Y+ GY KT YS DE SGG YG GG G T GGYG
Sbjct: 342 EGGYNKTS-YSSDEPASGGG-YEGGYKKTGYSGGIDETSGG-YGAGGGGYSDVTDGGYGK 512
Query: 117 GGGGGGEYSTGGGGGGY--STGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTA 174
GG Y GGGG Y +T GGY G DEVDYK+EEKHHK LEHLGELG +
Sbjct: 513 PSGG---YGAAGGGGVYPDTTTDGGYSKTSGGGYDDEVDYKKEEKHHKHLEHLGELGAAS 683
Query: 175 AGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGK 234
A AYALHEKHK+EKDPE+AHRHK+EEEVAAAAAVG+GGFAFHEHHEKKEAKE++EE+HGK
Sbjct: 684 AAAYALHEKHKAEKDPEHAHRHKIEEEVAAAAAVGSGGFAFHEHHEKKEAKEQDEEAHGK 863
Query: 235 KHHHLFG 241
KHHHLFG
Sbjct: 864 KHHHLFG 884
>TC224981 similar to UP|O50000 (O50000) Abscisic stress ripening protein
homolog, partial (50%)
Length = 849
Score = 280 bits (716), Expect = 4e-76
Identities = 149/245 (60%), Positives = 171/245 (68%), Gaps = 4/245 (1%)
Frame = +1
Query: 1 MAEENRDRGLFHHHKNED-RPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYE 59
+ E++ LFH HK+ED +PT+TETGY+ TSYS +PS DYDSG+ SYETSGG YE
Sbjct: 79 LEEKHHKHHLFHQHKDEDNKPTETETGYDNTSYS--KPSD-DYDSGFSNPSYETSGGAYE 249
Query: 60 TGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP--SGGGYGTGGAGGGYSTGGGYGTG 117
T G+NKT+YS+D+ SGGGY G GG G G
Sbjct: 250 T----------------------GHNKTSYSSDDQPASGGGYKKTGYSGGIDETSGGGGY 363
Query: 118 GGGGGEYSTGGGGGGYS-TGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAG 176
GGGGG YS GGY+ T GG YG DEVDYK+EEKHHK LEHLGELG AAG
Sbjct: 364 GGGGGVYSDTTTDGGYTRTSGGRYG--------DEVDYKKEEKHHKSLEHLGELGAAAAG 519
Query: 177 AYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKH 236
AYALHEKHK+EKDPE+AHRHK+EEEVAAAAAVG+GGFAFHEHHEKKEAKE++EE+HGKKH
Sbjct: 520 AYALHEKHKAEKDPEHAHRHKIEEEVAAAAAVGSGGFAFHEHHEKKEAKEQDEEAHGKKH 699
Query: 237 HHLFG 241
HHLFG
Sbjct: 700 HHLFG 714
>TC224571 similar to UP|Q9ZRB6 (Q9ZRB6) Ci21A protein, complete
Length = 762
Score = 168 bits (425), Expect = 2e-42
Identities = 74/91 (81%), Positives = 84/91 (91%)
Frame = +3
Query: 151 EVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGA 210
EVDYK+EEKHHK LEHLGELG AAGAYALHEKH+++KDPE+AHRHK+EEE+AAAA VGA
Sbjct: 168 EVDYKKEEKHHKHLEHLGELGAAAAGAYALHEKHEAKKDPEHAHRHKVEEEIAAAATVGA 347
Query: 211 GGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
GGF HEHHEKKE K+E+EE+HGKKHHHLFG
Sbjct: 348 GGFVLHEHHEKKEVKKEDEEAHGKKHHHLFG 440
>BF324843
Length = 313
Score = 147 bits (371), Expect = 4e-36
Identities = 70/111 (63%), Positives = 85/111 (76%), Gaps = 1/111 (0%)
Frame = +2
Query: 131 GGYS-TGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKD 189
GGYS T GGGY DEVDY +E H LEHLGEL +A AY+LH+KHK++KD
Sbjct: 5 GGYSKTSGGGYD--------DEVDYNKENNLHMHLEHLGELSAASAAAYSLHDKHKAQKD 160
Query: 190 PENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
PE+AH HK+ EEVAAAAAVG+ GFAFHEHH+K EA++++EE+H KKHHHLF
Sbjct: 161 PEHAHTHKI*EEVAAAAAVGSDGFAFHEHHDKNEAQDQDEEAHQKKHHHLF 313
>BE190928
Length = 235
Score = 109 bits (273), Expect = 1e-24
Identities = 53/77 (68%), Positives = 62/77 (79%)
Frame = -1
Query: 162 KKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEK 221
K LEHL LG + A ALH KHK+EKDPE+A R K+EE+VAAAAAVG+GGFAFH HH K
Sbjct: 235 KHLEHLS*LGPASVAASALHYKHKAEKDPEHALRPKIEEQVAAAAAVGSGGFAFHLHHAK 56
Query: 222 KEAKEEEEESHGKKHHH 238
K AKE++EE+HGKK HH
Sbjct: 55 K*AKEQDEEAHGKKLHH 5
>AI736478
Length = 419
Score = 102 bits (254), Expect = 2e-22
Identities = 43/54 (79%), Positives = 50/54 (91%)
Frame = +2
Query: 188 KDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
+DPE+AHRHK+EEE+AAAA VGAGGF HEHHEKKE K+E+EE+HGKKHHHLFG
Sbjct: 5 EDPEHAHRHKVEEEIAAAATVGAGGFVLHEHHEKKEVKKEDEEAHGKKHHHLFG 166
>TC224759 similar to UP|Q9SPF3 (Q9SPF3) Ripening induced protein (Fragment),
partial (78%)
Length = 427
Score = 74.3 bits (181), Expect = 5e-14
Identities = 34/56 (60%), Positives = 39/56 (68%)
Frame = +1
Query: 186 SEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
SE P+ E+ AA VGAGGF HEHHEKKE K+E+EE+HGKKHHHLFG
Sbjct: 175 SEIVPDTVVFDDFEKFPPTAATVGAGGFVLHEHHEKKEVKKEDEEAHGKKHHHLFG 342
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 73.2 bits (178), Expect = 1e-13
Identities = 48/122 (39%), Positives = 60/122 (48%), Gaps = 12/122 (9%)
Frame = -3
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G + ++GGG G + S+ + S GGG G ++ S GGG
Sbjct: 595 GGGYGGGGSQGGGGSAGGGGS*GGSGGSAGGGEGSGSQGGGSGGGGSQGGGSGS--GGGG 422
Query: 99 YGTGGAGG----GYSTGGGYGTGGGGGGEYSTGGGGGG--------YSTGGGGYGNDDSG 146
YG GG+GG G S GGG G GGGGGG Y GG GG Y G GG G+ D G
Sbjct: 421 YGGGGSGGSEGGGSSGGGGGGGGGGGGGGYGGGGDKGGGIGGDKGDYHHGKGGKGDKDGG 242
Query: 147 NR 148
++
Sbjct: 241 DK 236
Score = 69.7 bits (169), Expect = 1e-12
Identities = 51/133 (38%), Positives = 63/133 (47%), Gaps = 5/133 (3%)
Frame = -3
Query: 36 EPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNS-GGGGYDSGYNKTAYSTDEP 94
E GG G K GGGY G ++ S S GG G +G + + S
Sbjct: 637 EGGGGSAGGGGSKG-----GGGYGGGGSQGGGGSAGGGGS*GGSGGSAGGGEGSGSQGGG 473
Query: 95 SGGGY----GTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRD 150
SGGG G+G GGGY GG G+GG GG S GGGGGG GGGGYG G++
Sbjct: 472 SGGGGSQGGGSGSGGGGY---GGGGSGGSEGGGSSGGGGGGGGGGGGGGYGG--GGDKGG 308
Query: 151 EVDYKEEEKHHKK 163
+ + + HH K
Sbjct: 307 GIGGDKGDYHHGK 269
Score = 59.7 bits (143), Expect = 1e-09
Identities = 37/80 (46%), Positives = 41/80 (51%), Gaps = 8/80 (10%)
Frame = -3
Query: 75 SGGGGYDSGYNKTAYSTDEPSGGGYGTGG-AGGGYSTGGGYGTGGGGGGEYSTGGG---- 129
S GGG +G + GG G GG AGGG S GG G+ GGG G S GGG
Sbjct: 640 SEGGGGSAGGGGSKGGGGYGGGGSQGGGGSAGGGGS*GGSGGSAGGGEGSGSQGGGSGGG 461
Query: 130 ---GGGYSTGGGGYGNDDSG 146
GGG +GGGGYG SG
Sbjct: 460 GSQGGGSGSGGGGYGGGGSG 401
Score = 59.3 bits (142), Expect = 2e-09
Identities = 42/109 (38%), Positives = 48/109 (43%), Gaps = 7/109 (6%)
Frame = -3
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG G S+ G G + G + SGGGGY G + + GGG
Sbjct: 544 GGGS*GGSGGSAGGGEGSGSQGGGSGGGGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGG 365
Query: 99 YGTGGAGGG-YSTGGGYGTG-GGGGGEYSTGGGG-----GGYSTGGGGY 140
G GG GGG Y GG G G GG G+Y G GG GG GGGGY
Sbjct: 364 GGGGGGGGGGYGGGGDKGGGIGGDKGDYHHGKGGKGDKDGGDKGGGGGY 218
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 69.7 bits (169), Expect = 1e-12
Identities = 38/76 (50%), Positives = 40/76 (52%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G + E G G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 176 GGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGG--- 346
Query: 136 GGGGYGNDDSGNRRDE 151
GGGG G G RR+E
Sbjct: 347 GGGGGGGGGRGGRREE 394
Score = 69.3 bits (168), Expect = 1e-12
Identities = 42/110 (38%), Positives = 45/110 (40%)
Frame = +3
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG G + GGG G GGGG G + GG
Sbjct: 66 GGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGREXXGG 245
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNR 148
G GG GGG GGG G GGGGG GGGGGG GGGG G + G R
Sbjct: 246 GGEGGGGGGGGGGGGGGGXGGGGGGRGEGGGGGGGGGGGGGGGGEGGGRR 395
Score = 65.1 bits (157), Expect = 3e-11
Identities = 37/74 (50%), Positives = 37/74 (50%)
Frame = +3
Query: 73 PNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG 132
P GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 18 PGGGGGGGGGG------------GGGGGGGGGGGGGEGGGGGGGGGGGGG----GGGGGG 149
Query: 133 YSTGGGGYGNDDSG 146
GGGG G G
Sbjct: 150 GGRGGGGGGGGGGG 191
Score = 65.1 bits (157), Expect = 3e-11
Identities = 41/94 (43%), Positives = 43/94 (45%), Gaps = 2/94 (2%)
Frame = +3
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
GGG G + GGGG G+ E GGG GG GGG GGG
Sbjct: 129 GGGGGGGGGRGGGGGGGGGGGGGGGGGGGF----LGGREXXGGGGEGGGGGGGGGGGGGG 296
Query: 115 GTGGGGGG--EYSTGGGGGGYSTGGGGYGNDDSG 146
G GGGGGG E GGGGGG GGGG G G
Sbjct: 297 GXGGGGGGRGEGGGGGGGGGGGGGGGGEGGGRRG 398
Score = 64.7 bits (156), Expect = 4e-11
Identities = 35/66 (53%), Positives = 35/66 (53%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 38 GGGGGGGG-----------GGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGG 184
Query: 136 GGGGYG 141
GGGG G
Sbjct: 185 GGGGGG 202
Score = 64.3 bits (155), Expect = 5e-11
Identities = 34/69 (49%), Positives = 36/69 (51%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G + E GGG G GG GGG GGG G G GGGG GGGGGG
Sbjct: 201 GGGGGFLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGGGRGEGGGGGGGGGGGGGGGGE 380
Query: 136 GGGGYGNDD 144
GGG G +
Sbjct: 381 GGGRRGGGE 407
Score = 64.3 bits (155), Expect = 5e-11
Identities = 31/51 (60%), Positives = 31/51 (60%)
Frame = +2
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GG GGG GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 41 GGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGG 193
Score = 63.9 bits (154), Expect = 6e-11
Identities = 36/77 (46%), Positives = 37/77 (47%), Gaps = 6/77 (7%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG--- 132
GGGG G + GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 42 GGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFL 221
Query: 133 ---YSTGGGGYGNDDSG 146
GGGG G G
Sbjct: 222 GGREXXGGGGEGGGGGG 272
Score = 63.5 bits (153), Expect = 8e-11
Identities = 34/65 (52%), Positives = 35/65 (53%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYST 135
GGGG G GGG G GG GGG GGG G GGGGGG GGGGGG
Sbjct: 36 GGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGG----GGGGGGGGG 203
Query: 136 GGGGY 140
GGGG+
Sbjct: 204 GGGGF 218
Score = 63.5 bits (153), Expect = 8e-11
Identities = 38/79 (48%), Positives = 38/79 (48%)
Frame = +1
Query: 73 PNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGG 132
P GGGG G GGG G GG GGG GGG G GGGGG GGGGGG
Sbjct: 19 PGGGGGGGGGGGGGGGGG----GGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGG 186
Query: 133 YSTGGGGYGNDDSGNRRDE 151
GGGG G G R E
Sbjct: 187 GGGGGGGGGVF*GGERXRE 243
Score = 63.2 bits (152), Expect = 1e-10
Identities = 39/95 (41%), Positives = 43/95 (45%), Gaps = 3/95 (3%)
Frame = +2
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGY---NKTAYSTDEPSGGGYGTGGAGGGYSTG 111
GGG G K GGGG G+ + GGG G GG GGG G
Sbjct: 128 GGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGG 307
Query: 112 GGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GG G GGGGGG GGGGGG GG G ++ G
Sbjct: 308 GGGGEGGGGGG----GGGGGGGGGGGRGGRREEGG 400
Score = 62.4 bits (150), Expect = 2e-10
Identities = 43/120 (35%), Positives = 45/120 (36%), Gaps = 12/120 (10%)
Frame = +1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG+ G GGG G + GGGG G GGG
Sbjct: 4 GGEVCPGGGGGGGGGGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGGG 183
Query: 99 YGTGGAGG------------GYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
G GG GG G GGG G GGGGGG GGGGGG GGGG G G
Sbjct: 184 GGGGGGGGGGVF*GGERXREGGERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGGGGGG 363
Score = 62.4 bits (150), Expect = 2e-10
Identities = 46/130 (35%), Positives = 46/130 (35%), Gaps = 15/130 (11%)
Frame = +1
Query: 37 PSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
P GG G GGG G E GGGG G G
Sbjct: 19 PGGGGGGGG--------GGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGG 174
Query: 97 GGYGTGGAGGGYST---------------GGGYGTGGGGGGEYSTGGGGGGYSTGGGGYG 141
GG G GG GGG GGG G GGGGGG GGGGGG GGGG G
Sbjct: 175 GGGGGGGGGGGGGVF*GGERXREGGERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGGG 354
Query: 142 NDDSGNRRDE 151
G E
Sbjct: 355 GGGGGGEGRE 384
Score = 62.0 bits (149), Expect = 2e-10
Identities = 38/90 (42%), Positives = 39/90 (43%), Gaps = 14/90 (15%)
Frame = +3
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGG-------------- 121
GGGG G + GGG G GG GGG GGG G GGGGG
Sbjct: 60 GGGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGREXX 239
Query: 122 GEYSTGGGGGGYSTGGGGYGNDDSGNRRDE 151
G GGGGGG GGGG G G R E
Sbjct: 240 GGGGEGGGGGGGGGGGGGGGXGGGGGGRGE 329
Score = 61.6 bits (148), Expect = 3e-10
Identities = 30/52 (57%), Positives = 30/52 (57%)
Frame = +3
Query: 97 GGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNR 148
GG G G GGG GGG G GGGGGG GGGGGG GGGG G G R
Sbjct: 3 GGRGLPGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGR 158
Score = 60.5 bits (145), Expect = 7e-10
Identities = 38/93 (40%), Positives = 39/93 (41%), Gaps = 11/93 (11%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGG-----------YSTGGGYGTGGGGGGEY 124
GGGG G GGG G GG GGG GGG G GGGGGG
Sbjct: 116 GGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGG 295
Query: 125 STGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEE 157
GGGGGG GGGG G G +EE
Sbjct: 296 GGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREE 394
Score = 57.4 bits (137), Expect = 6e-09
Identities = 39/101 (38%), Positives = 40/101 (38%)
Frame = +2
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG G K GGG G + E G GG G GGG
Sbjct: 128 GGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGG--------GGGG 283
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
G GG GGG GG G GGGGGG GG GG GGGG
Sbjct: 284 GGGGGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREEGGGG 406
Score = 55.5 bits (132), Expect = 2e-08
Identities = 28/51 (54%), Positives = 28/51 (54%)
Frame = +1
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGG G GGG GGG G GGGGGG GG GGG GGGG G G
Sbjct: 1 GGGEVCPGGGGGGGGGGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGG 153
Score = 52.0 bits (123), Expect = 2e-07
Identities = 27/53 (50%), Positives = 29/53 (53%)
Frame = +2
Query: 96 GGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNR 148
G + GG GGG GGG G GGGGGG GGG GG GGGG G G +
Sbjct: 5 GERFARGGGGGG--GGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGK 157
Score = 34.7 bits (78), Expect = 0.040
Identities = 19/38 (50%), Positives = 19/38 (50%)
Frame = +2
Query: 112 GGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRR 149
GG GGGG GGGGGG GGGG G G R
Sbjct: 2 GGERFARGGGG----GGGGGGGGGGGGGGGGGGGGGGR 103
Score = 33.9 bits (76), Expect = 0.068
Identities = 18/34 (52%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Frame = +1
Query: 120 GGGEYSTGGGGGG--YSTGGGGYGNDDSGNRRDE 151
GGGE GGGGGG GGGG G G R E
Sbjct: 1 GGGEVCPGGGGGGGGGGGGGGGGGGGGGGGRGGE 102
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 69.3 bits (168), Expect = 1e-12
Identities = 39/84 (46%), Positives = 44/84 (51%), Gaps = 3/84 (3%)
Frame = +1
Query: 66 SSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYS--TGGGYGTGG-GGGG 122
S S + G G + + + GGG G GG GGGY GGGYG GG GGGG
Sbjct: 238 SEQSMKDAIEGMNGQNLDGRNITVNEAQSRGGGGGGGGGGGGYGGGRGGGYGGGGRGGGG 417
Query: 123 EYSTGGGGGGYSTGGGGYGNDDSG 146
Y+ GGGGGY GGGGYG G
Sbjct: 418 GYNRSGGGGGYG-GGGGYGGGGGG 486
Score = 67.0 bits (162), Expect = 7e-12
Identities = 46/132 (34%), Positives = 56/132 (41%), Gaps = 10/132 (7%)
Frame = +1
Query: 18 DRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGG 77
DR T G+ ++++++ K + E G G N T + + GG
Sbjct: 190 DRETGRSRGFGFVTFASEQSM---------KDAIEGMNGQNLDGRNITVNEAQSRGGGGG 342
Query: 78 GG-----YDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG--GGEYSTGGGG 130
GG Y G GGGY G GGGY GGGYG GGGG GG G GG
Sbjct: 343 GGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYGGGGGYGGGGGGGYGGGRDRGYGG 522
Query: 131 G---GYSTGGGG 139
G GYS GG G
Sbjct: 523 GGDRGYSRGGDG 558
Score = 59.3 bits (142), Expect = 2e-09
Identities = 38/88 (43%), Positives = 42/88 (47%)
Frame = +1
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
GGGY G + Y GG G GYN++ GGGYG GG GGGY G
Sbjct: 355 GGGY--GGGRGGGYG-----GGGRGGGGGYNRSGGGGGYGGGGGYG-GGGGGGYGGGRDR 510
Query: 115 GTGGGGGGEYSTGGGGGGYSTGGGGYGN 142
G GGGG YS GG G G GG+ N
Sbjct: 511 GYGGGGDRGYSRGGDG-----GDGGWRN 579
Score = 31.6 bits (70), Expect = 0.34
Identities = 25/76 (32%), Positives = 29/76 (37%), Gaps = 2/76 (2%)
Frame = +1
Query: 39 GGDYDSGYKKSS--YETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
GG Y G + Y SGGG GY Y GGGGY G ++ Y G
Sbjct: 379 GGGYGGGGRGGGGGYNRSGGG--GGYGGGGGYG----GGGGGGYGGGRDR-GYGGGGDRG 537
Query: 97 GGYGTGGAGGGYSTGG 112
G G GG+ G
Sbjct: 538 YSRGGDGGDGGWRN*G 585
>TC214257 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (7%)
Length = 607
Score = 68.2 bits (165), Expect = 3e-12
Identities = 39/82 (47%), Positives = 42/82 (50%), Gaps = 7/82 (8%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGG--GGGGEYSTGGGGGGY 133
GGGGY G + E GGGY G GGGY GGGYG GG GGGG Y GG GY
Sbjct: 8 GGGGYGGGGRR------EGGGGGYNRNGGGGGYGGGGGYGGGGGHGGGGGYGGGGRDRGY 169
Query: 134 STGGG-----GYGNDDSGNRRD 150
GG G G D G+ R+
Sbjct: 170 GGDGGSRYSRGGGASDGGSWRN 235
Score = 46.6 bits (109), Expect = 1e-05
Identities = 38/111 (34%), Positives = 44/111 (39%), Gaps = 5/111 (4%)
Frame = +2
Query: 39 GGDYDSGYKKSSYETSGGGYET-----GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDE 93
GG Y G ++ E GGGY GY Y GGGG+
Sbjct: 11 GGGYGGGGRR---EGGGGGYNRNGGGGGYGGGGGYG------GGGGHG------------ 127
Query: 94 PSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDD 144
GGGYG GG GY GG GG YS GGG ++ GG + N D
Sbjct: 128 -GGGGYGGGGRDRGY--------GGDGGSRYSRGGG----ASDGGSWRN*D 241
>AI431095 weakly similar to PIR|T09789|T097 abscisic acid- and
water-stress-inducible protein LP3-1 - loblolly pine,
partial (30%)
Length = 115
Score = 66.2 bits (160), Expect = 1e-11
Identities = 29/38 (76%), Positives = 33/38 (86%)
Frame = -1
Query: 202 VAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHL 239
VAAAA VG GGFA HEHH+KK AKE++EE+HGKK HHL
Sbjct: 115 VAAAAQVGYGGFALHEHHDKKIAKEQDEEAHGKKQHHL 2
>TC215791 similar to GB|CAE76446.1|38567152|BX842633 related to peroxisomal
membrane protein pex13 {Neurospora crassa;} , partial
(9%)
Length = 1440
Score = 65.1 bits (157), Expect = 3e-11
Identities = 46/130 (35%), Positives = 60/130 (45%), Gaps = 3/130 (2%)
Frame = +3
Query: 21 TDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGY 80
+D N+ + P+ ++ Y S+Y GGY + N S Y + S GG
Sbjct: 279 SDRSAAVNRNTLGRPVPTR-PWEQNYGNSTY----GGYGSTMNYNSGYGSGMYGSSYGGL 443
Query: 81 DSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGG-GGGEYSTGGGG--GGYSTGG 137
G ++Y GG YG GGY GG YG+ G GGG Y++G GG GGY GG
Sbjct: 444 GGGMYGSSYG-----GGMYGNSMYRGGY--GGMYGSSGMYGGGMYNSGLGGPMGGYGMGG 602
Query: 138 GGYGNDDSGN 147
G YG D N
Sbjct: 603 GPYGEQDPNN 632
>TC214360 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding protein,
partial (44%)
Length = 630
Score = 64.7 bits (156), Expect = 4e-11
Identities = 43/97 (44%), Positives = 49/97 (50%), Gaps = 1/97 (1%)
Frame = +2
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
GGGY G Y GGG + GYN+ +GGG G GG GGGY GGGY
Sbjct: 5 GGGYSRG---GGGYG-----GGGGRREGGYNR--------NGGGGGYGGGGGGYGGGGGY 136
Query: 115 GTGGGGGGEYSTGGGGGG-YSTGGGGYGNDDSGNRRD 150
GGGG + GG GG YS GGGG D G+ R+
Sbjct: 137 ---GGGGRDRGYGGDGGSRYSRGGGG---SDGGSWRN 229
Score = 61.6 bits (148), Expect = 3e-10
Identities = 44/109 (40%), Positives = 47/109 (42%)
Frame = +2
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GG Y G Y GG E GYN+ N GGGGY G GGG
Sbjct: 5 GGGYSRG--GGGYGGGGGRREGGYNR---------NGGGGGYGGG------------GGG 115
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
YG GG GG GY GG GG YS GGGG + GG + N D N
Sbjct: 116 YGGGGGYGGGGRDRGY--GGDGGSRYSRGGGG----SDGGSWRN*DYVN 244
Score = 50.1 bits (118), Expect = 9e-07
Identities = 34/67 (50%), Positives = 34/67 (50%), Gaps = 1/67 (1%)
Frame = +2
Query: 76 GGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYS- 134
GGGGY G GGGYG GGG GGY GGGGG GGGGGG
Sbjct: 2 GGGGYSRG------------GGGYG----GGGGRREGGYNRNGGGGGY---GGGGGG--Y 118
Query: 135 TGGGGYG 141
GGGGYG
Sbjct: 119 GGGGGYG 139
Score = 34.3 bits (77), Expect = 0.052
Identities = 19/38 (50%), Positives = 20/38 (52%), Gaps = 11/38 (28%)
Frame = +2
Query: 120 GGGEYSTGGGG---------GGYST--GGGGYGNDDSG 146
GGG YS GGGG GGY+ GGGGYG G
Sbjct: 2 GGGGYSRGGGGYGGGGGRREGGYNRNGGGGGYGGGGGG 115
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 64.3 bits (155), Expect = 5e-11
Identities = 47/114 (41%), Positives = 52/114 (45%), Gaps = 13/114 (11%)
Frame = -1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDS---GYNKTAYSTDEPS 95
GGD D G + GGG E S+ GGGG DS G + +
Sbjct: 321 GGDGDGGGW*GAECGDGGGGEC--------SSQGGGGGGGGGDSIQGGGDGGGGGGE*TG 166
Query: 96 GGGYGTGGAGGGYSTG----------GGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
GGG TGG GGG G GG G GGGGGGEY GG GG Y+ GGGG
Sbjct: 165 GGGE*TGGGGGGDK*G*TGGGGGGECGGGGDGGGGGGEYVGGGAGGEYTGGGGG 4
Score = 55.1 bits (131), Expect = 3e-08
Identities = 40/101 (39%), Positives = 44/101 (42%), Gaps = 12/101 (11%)
Frame = -1
Query: 43 DSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYG-- 100
D G + S + GGG G D GG G G T + GGG G
Sbjct: 276 DGGGGECSSQGGGGGGGGG---------DSIQGGGDGGGGGGE*TGGGGE*TGGGGGGDK 124
Query: 101 ---TGGAGGGYSTGGGYGTGGGG-------GGEYSTGGGGG 131
TGG GGG GGG G GGGG GGEY+ GGGGG
Sbjct: 123 *G*TGGGGGGECGGGGDGGGGGGEYVGGGAGGEYTGGGGGG 1
Score = 51.2 bits (121), Expect = 4e-07
Identities = 47/116 (40%), Positives = 48/116 (40%), Gaps = 8/116 (6%)
Frame = -1
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGG 98
GGD G Y GGG G + E GGGG E S G
Sbjct: 366 GGDGGGG---ELY*IGGGG---GDGDGGGW*GAECGDGGGG-------------ECSSQG 244
Query: 99 YGTGGAGGGYSTGGGYGTGGGG-----GGEYSTGGGGGGYS---TGGGGYGNDDSG 146
G GG GG GGG G GGGG GGE TGGGGGG TGGGG G G
Sbjct: 243 GGGGGGGGDSIQGGGDGGGGGGE*TGGGGE*-TGGGGGGDK*G*TGGGGGGECGGG 79
Score = 49.7 bits (117), Expect = 1e-06
Identities = 42/119 (35%), Positives = 50/119 (41%), Gaps = 19/119 (15%)
Frame = -1
Query: 47 KKSSYETS--GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGG- 103
+KSS + GGG Y + +G GG D G GGG G GG
Sbjct: 522 EKSSQKNQ*KGGGGGDAYETPMTGGKGPSYTGAGGGDDGGGGEL***TGVGGGGDGGGGE 343
Query: 104 -----AGGGYSTGGGY---GTGGGGGGEYSTGGGGGGYSTG--------GGGYGNDDSG 146
GGG GGG+ G GGGGE S+ GGGGG G GGG G + +G
Sbjct: 342 LY*IGGGGGDGDGGGW*GAECGDGGGGECSSQGGGGGGGGGDSIQGGGDGGGGGGE*TG 166
Score = 45.4 bits (106), Expect = 2e-05
Identities = 47/125 (37%), Positives = 58/125 (45%), Gaps = 21/125 (16%)
Frame = -1
Query: 39 GGD-YDS---GYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKT---AYST 91
GGD Y++ G K SY +GGG + G + + GGG D G +
Sbjct: 483 GGDAYETPMTGGKGPSYTGAGGGDDGGGGEL***T-----GVGGGGDGGGGELY*IGGGG 319
Query: 92 DEPSGGGY-----GTGGAGGGYSTGGGYGTGGG----GGGEYSTGG-----GGGGYSTGG 137
+ GGG+ G GG G S GGG G GGG GGG+ GG GGGG TGG
Sbjct: 318 GDGDGGGW*GAECGDGGGGECSSQGGGGGGGGGDSIQGGGD-GGGGGGE*TGGGGE*TGG 142
Query: 138 GGYGN 142
GG G+
Sbjct: 141 GGGGD 127
>TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich protein,
partial (37%)
Length = 750
Score = 61.6 bits (148), Expect = 3e-10
Identities = 43/113 (38%), Positives = 45/113 (39%), Gaps = 9/113 (7%)
Frame = -1
Query: 39 GGDYD--SGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
GGD G S GGY G + EP SG G GY P G
Sbjct: 630 GGDAGPGGGGPGGSAGPGSGGYGGGPGGDTGPGGGEPGSGAGPGSGGYGGGPGGDAGPGG 451
Query: 97 GGYGTGGAGGGYSTG-------GGYGTGGGGGGEYSTGGGGGGYSTGGGGYGN 142
GG G G GG G GGYG G GGG GG GGG GGGG G+
Sbjct: 450 GGPGGGAGPGGGEPGSGAGPGSGGYGGGPGGGVGPGGGGPGGGAGRGGGGPGS 292
Score = 60.8 bits (146), Expect = 5e-10
Identities = 45/127 (35%), Positives = 48/127 (37%), Gaps = 6/127 (4%)
Frame = -1
Query: 26 GYNKTSYSTDEPSGGDYDSGYKKSSYETSGG-GYETGYNKT-----SSYSTDEPNSGGGG 79
GY P GG+ SG S GG G + G + EP SG G
Sbjct: 570 GYGGGPGGDTGPGGGEPGSGAGPGSGGYGGGPGGDAGPGGGGPGGGAGPGGGEPGSGAGP 391
Query: 80 YDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGG 139
GY P GGG G G GG G G G GGG G S GG GG GG G
Sbjct: 390 GSGGYGGGPGGGVGPGGGGPGGGAGRGGGGPGSGAGPSGGGPGGGSYGGEPGGGGPGGDG 211
Query: 140 YGNDDSG 146
YG G
Sbjct: 210 YGGGPGG 190
Score = 58.2 bits (139), Expect = 3e-09
Identities = 47/119 (39%), Positives = 50/119 (41%), Gaps = 9/119 (7%)
Frame = -1
Query: 37 PSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNS----GGGGYDSGYN-KTAYST 91
P GG Y G + GG Y G + P G GGY G T
Sbjct: 705 PVGGGYGGG-PGGGAGSGGGSYGGGPGGDAGPGGGGPGGSAGPGSGGYGGGPGGDTGPGG 529
Query: 92 DEP-SGGGYGTGGAGGGYSTGGGYGTGGGG-GGEYSTGGG--GGGYSTGGGGYGNDDSG 146
EP SG G G+GG GGG GG G GGGG GG GGG G G G GGYG G
Sbjct: 528 GEPGSGAGPGSGGYGGG--PGGDAGPGGGGPGGGAGPGGGEPGSGAGPGSGGYGGGPGG 358
Score = 56.6 bits (135), Expect = 1e-08
Identities = 27/54 (50%), Positives = 28/54 (51%)
Frame = -1
Query: 94 PSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
P GGGYG G GG S GG YG G GG GG GG G GGYG G+
Sbjct: 705 PVGGGYGGGPGGGAGSGGGSYGGGPGGDAGPGGGGPGGSAGPGSGGYGGGPGGD 544
Score = 52.0 bits (123), Expect = 2e-07
Identities = 45/115 (39%), Positives = 46/115 (39%), Gaps = 5/115 (4%)
Frame = -1
Query: 37 PSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
P GG+ SG S GGG G GGGG SG S P G
Sbjct: 426 PGGGEPGSGAGPGS-GGYGGGPGGGVGPGGGGPGGGAGRGGGGPGSGAGP---SGGGPGG 259
Query: 97 GGYGT---GGAGGGYSTGGGYGTGGG--GGGEYSTGGGGGGYSTGGGGYGNDDSG 146
G YG GG GG GGG G G G GGG G GGG Y GGYG G
Sbjct: 258 GSYGGEPGGGGPGGDGYGGGPGGGAGPDGGGPKDGGFGGGPY----GGYGPRSHG 106
Score = 50.8 bits (120), Expect = 5e-07
Identities = 35/83 (42%), Positives = 37/83 (44%), Gaps = 12/83 (14%)
Frame = -1
Query: 77 GGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGG----------YGTGGGGGGEYST 126
GGGY G A S GG YG GG GG GGG G GGG GG+
Sbjct: 699 GGGYGGGPGGGAGS----GGGSYG-GGPGGDAGPGGGGPGGSAGPGSGGYGGGPGGDTGP 535
Query: 127 GGG--GGGYSTGGGGYGNDDSGN 147
GGG G G G GGYG G+
Sbjct: 534 GGGEPGSGAGPGSGGYGGGPGGD 466
Score = 48.9 bits (115), Expect = 2e-06
Identities = 29/52 (55%), Positives = 29/52 (55%), Gaps = 6/52 (11%)
Frame = -1
Query: 96 GGGYGTG-GAGGGYSTGGGYGTG-----GGGGGEYSTGGGGGGYSTGGGGYG 141
GGG G G G GGG GGGYG G G GGG Y GG GG GGGG G
Sbjct: 747 GGGPGGGVGPGGGGPVGGGYGGGPGGGAGSGGGSYG-GGPGGDAGPGGGGPG 595
Score = 46.6 bits (109), Expect = 1e-05
Identities = 25/49 (51%), Positives = 27/49 (55%)
Frame = -1
Query: 99 YGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGN 147
YG GG GGG GGG GGG G GG GGG +GGG YG G+
Sbjct: 750 YG-GGPGGGVGPGGGGPVGGGYG-----GGPGGGAGSGGGSYGGGPGGD 622
Score = 37.4 bits (85), Expect = 0.006
Identities = 29/86 (33%), Positives = 31/86 (35%)
Frame = -1
Query: 37 PSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSG 96
P GG G + G +G EP GG G D GY P G
Sbjct: 348 PGGGGPGGGAGRGGGGPGSGAGPSGGGPGGGSYGGEPGGGGPGGD-GYGGGPGGGAGPDG 172
Query: 97 GGYGTGGAGGGYSTGGGYGTGGGGGG 122
GG GG GGG GGYG G G
Sbjct: 171 GGPKDGGFGGG--PYGGYGPRSHGFG 100
>TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protein [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(88%)
Length = 1085
Score = 61.6 bits (148), Expect = 3e-10
Identities = 41/107 (38%), Positives = 50/107 (46%), Gaps = 4/107 (3%)
Frame = +2
Query: 39 GGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEP-SGG 97
GG SG + SG G + + SS ++ +SG GG +G +Y+ SG
Sbjct: 518 GGGSGSGAGAGAGSGSGSGSGSSSSSASSSASSSSSSGSGGSGAGSEAGSYAGSRAGSGS 697
Query: 98 GYGTGGAGGGYSTGGGYGTGGGGGGEYSTG---GGGGGYSTGGGGYG 141
G G GGG GGG G GGGGG Y G G G GY GGG G
Sbjct: 698 GRSQGRGGGG---GGGGGGGGGGGSGYGHGEGYGHGEGYGQGGGD*G 829
Score = 55.5 bits (132), Expect = 2e-08
Identities = 34/95 (35%), Positives = 44/95 (45%), Gaps = 5/95 (5%)
Frame = +2
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYS----- 109
GGG +G + + SG G S + +A S+ GG G G G Y+
Sbjct: 518 GGGSGSGAGAGAGSGS---GSGSGSSSSSASSSASSSSSSGSGGSGAGSEAGSYAGSRAG 688
Query: 110 TGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDD 144
+G G G GGGG GGGGGG GG GYG+ +
Sbjct: 689 SGSGRSQGRGGGG----GGGGGGGGGGGSGYGHGE 781
Score = 51.2 bits (121), Expect = 4e-07
Identities = 32/94 (34%), Positives = 40/94 (42%), Gaps = 1/94 (1%)
Frame = +2
Query: 55 GGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGY 114
G G G S G G SG + ++ S+ S G+GG+G G G
Sbjct: 494 GLGLGLGIGGGSGSGAGAGAGSGSGSGSGSSSSSASSSASSSSSSGSGGSGAGSEAGSYA 673
Query: 115 GTGGGGGGEYSTG-GGGGGYSTGGGGYGNDDSGN 147
G+ G G S G GGGGG GGGG G G+
Sbjct: 674 GSRAGSGSGRSQGRGGGGGGGGGGGGGGGSGYGH 775
Score = 41.6 bits (96), Expect = 3e-04
Identities = 31/99 (31%), Positives = 38/99 (38%), Gaps = 1/99 (1%)
Frame = +2
Query: 25 TGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGY 84
+G +S S + SG S + G Y + S + GGGG G
Sbjct: 569 SGSGSSSSSASSSASSSSSSGSGGSGAGSEAGSYAGSRAGSGSGRSQGRGGGGGGGGGG- 745
Query: 85 NKTAYSTDEPSGGGYGTG-GAGGGYSTGGGYGTGGGGGG 122
GGG G+G G G GY G GYG GGG G
Sbjct: 746 -----------GGGGGSGYGHGEGYGHGEGYGQGGGD*G 829
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 61.2 bits (147), Expect = 4e-10
Identities = 42/94 (44%), Positives = 43/94 (45%), Gaps = 22/94 (23%)
Frame = +3
Query: 76 GGGG-----YDSGYNKTAYSTDEPSG-GGYGTGGAG---GGYSTGGGYGTGGGGGGE--Y 124
GGGG Y G + Y G GGYG GG G GGY GGGYG GGGGGG Y
Sbjct: 57 GGGGXXRGSYGXGGGRGGYGGGGRGGRGGYGGGGGGYGGGGYGGGGGYGGGGGGGGGGCY 236
Query: 125 STG-----------GGGGGYSTGGGGYGNDDSGN 147
G GGGGG GGGG G N
Sbjct: 237 KCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYN 338
Score = 61.2 bits (147), Expect = 4e-10
Identities = 44/115 (38%), Positives = 47/115 (40%), Gaps = 14/115 (12%)
Frame = +3
Query: 40 GDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGY 99
G Y G + Y G G GY GGGGY G Y GGGY
Sbjct: 78 GSYGXGGGRGGYGGGGRGGRGGYG-----------GGGGGYGGG----GYG----GGGGY 200
Query: 100 GTGGAGGGYSTGGGY------------GTGGGGGGEYSTGGGGGG--YSTGGGGY 140
G GG GGG GG Y GGGGGG Y GGGGGG Y+ G G+
Sbjct: 201 GGGGGGGG---GGCYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGH 356
Score = 48.9 bits (115), Expect = 2e-06
Identities = 26/47 (55%), Positives = 26/47 (55%), Gaps = 2/47 (4%)
Frame = +3
Query: 102 GGAGGGYSTGGGYGTGGGGGGEYSTG--GGGGGYSTGGGGYGNDDSG 146
G GGG G YG GGG GG Y G GG GGY GGGGYG G
Sbjct: 48 GTRGGGGXXRGSYGXGGGRGG-YGGGGRGGRGGYGGGGGGYGGGGYG 185
>TC215890 similar to PIR|F96770|F96770 protein RNA-binding protein F1O17.10
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (17%)
Length = 913
Score = 61.2 bits (147), Expect = 4e-10
Identities = 42/117 (35%), Positives = 51/117 (42%)
Frame = +2
Query: 29 KTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTA 88
+ +Y+ + P G Y G S GGGYE GG Y GY
Sbjct: 83 RVNYANERPRG--YGGGGFGSYGAVGGGGYE----------------GGSSYRGGYGGDN 208
Query: 89 YSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDS 145
YS ++ SG GYG GG Y +GG YG G G YS GGY+ GG GN +S
Sbjct: 209 YSRNDGSGYGYG----GGRYGSGGNYG-DSGSGNNYS-----GGYAGNAGGVGNHES 349
Score = 50.8 bits (120), Expect = 5e-07
Identities = 32/69 (46%), Positives = 37/69 (53%), Gaps = 10/69 (14%)
Frame = +2
Query: 89 YSTDEP---SGGGYGTGGA--GGGYSTGGGYGTGGGGGGEYSTGGGGG-GYSTG----GG 138
Y+ + P GGG+G+ GA GGGY G Y GG GG YS G G GY G GG
Sbjct: 92 YANERPRGYGGGGFGSYGAVGGGGYEGGSSY-RGGYGGDNYSRNDGSGYGYGGGRYGSGG 268
Query: 139 GYGNDDSGN 147
YG+ SGN
Sbjct: 269 NYGDSGSGN 295
>TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wall structural
protein 1.0 precursor (GRP 1.0), partial (27%)
Length = 447
Score = 61.2 bits (147), Expect = 4e-10
Identities = 31/64 (48%), Positives = 33/64 (51%)
Frame = +3
Query: 77 GGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTG 136
GGGY G + +GGGYG G GGG GGG G GG GG Y GGG G
Sbjct: 30 GGGYGGGEG-----AGQGAGGGYGGGAVGGGGGGGGGSGGGGARGGGYGAGGGSGEGGGH 194
Query: 137 GGGY 140
GGGY
Sbjct: 195 GGGY 206
Score = 57.8 bits (138), Expect = 4e-09
Identities = 34/58 (58%), Positives = 34/58 (58%), Gaps = 7/58 (12%)
Frame = +3
Query: 96 GGGYGTGGAGGGYSTGGGYGT----GGGGGGEYSTGGG--GGGYSTGGG-GYGNDDSG 146
GGGYG GG G G GGGYG GGGGGG S GGG GGGY GGG G G G
Sbjct: 30 GGGYG-GGEGAGQGAGGGYGGGAVGGGGGGGGGSGGGGARGGGYGAGGGSGEGGGHGG 200
Score = 47.8 bits (112), Expect = 5e-06
Identities = 27/45 (60%), Positives = 28/45 (62%), Gaps = 5/45 (11%)
Frame = +3
Query: 100 GTGGA-GGGYSTGGGYGTGGGGGGEYS----TGGGGGGYSTGGGG 139
G GGA GGGY GGG G G G GG Y GGGGGG +GGGG
Sbjct: 12 GAGGAHGGGY--GGGEGAGQGAGGGYGGGAVGGGGGGGGGSGGGG 140
Score = 46.6 bits (109), Expect = 1e-05
Identities = 25/45 (55%), Positives = 28/45 (61%)
Frame = +3
Query: 102 GGAGGGYSTGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSG 146
GGAGG + GGGYG GG G G+ + GG GGG GGGG G G
Sbjct: 9 GGAGGAH--GGGYG-GGEGAGQGAGGGYGGGAVGGGGGGGGGSGG 134
Score = 36.2 bits (82), Expect = 0.014
Identities = 22/43 (51%), Positives = 24/43 (55%), Gaps = 6/43 (13%)
Frame = +3
Query: 110 TGGGYGT---GGGGGGEYSTGGGGGGY---STGGGGYGNDDSG 146
T GG G GG GGGE + G GGGY + GGGG G SG
Sbjct: 3 TRGGAGGAHGGGYGGGEGAGQGAGGGYGGGAVGGGGGGGGGSG 131
Score = 34.7 bits (78), Expect = 0.040
Identities = 24/72 (33%), Positives = 27/72 (37%), Gaps = 1/72 (1%)
Frame = +3
Query: 38 SGGDYDSGYKKSSYETSGGGYETGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGG 97
+GG + GY GGG G Y GGGG A +GG
Sbjct: 15 AGGAHGGGY--------GGGEGAGQGAGGGYGGGAVGGGGGGGGGSGGGGARGGGYGAGG 170
Query: 98 GYGTGGA-GGGY 108
G G GG GGGY
Sbjct: 171 GSGEGGGHGGGY 206
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.303 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,949,270
Number of Sequences: 63676
Number of extensions: 194272
Number of successful extensions: 17275
Number of sequences better than 10.0: 1130
Number of HSP's better than 10.0 without gapping: 3220
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7919
length of query: 243
length of database: 12,639,632
effective HSP length: 95
effective length of query: 148
effective length of database: 6,590,412
effective search space: 975380976
effective search space used: 975380976
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0052b.4


