

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0052a.3
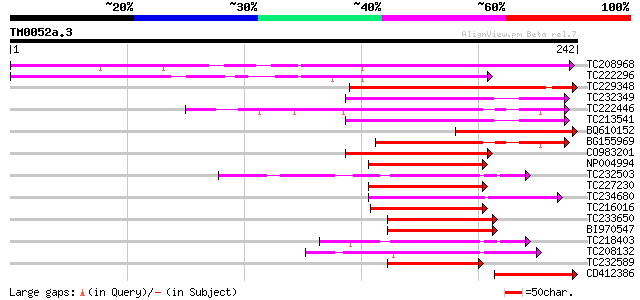
(242 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC208968 similar to UP|GBF4_ARATH (P42777) G-box binding factor ... 135 2e-32
TC222296 similar to UP|P93426 (P93426) Leucine zipper protein, p... 94 4e-20
TC229348 homologue to UP|Q94KA6 (Q94KA6) BZIP transcription fact... 78 3e-15
TC232349 similar to UP|O81241 (O81241) Dc3 promoter-binding fact... 71 5e-13
TC222446 similar to UP|O81241 (O81241) Dc3 promoter-binding fact... 69 2e-12
TC213541 similar to UP|Q9LES3 (Q9LES3) Promoter-binding factor-l... 65 2e-11
BQ610152 64 6e-11
BG155969 similar to PIR|T12622|T126 Dc3 promoter-binding factor-... 64 6e-11
CO983201 61 4e-10
NP004994 G/HBF-1 53 1e-07
TC232503 UP|Q39896 (Q39896) TGACG-motif-binding factor, partial ... 52 2e-07
TC227230 similar to UP|P93839 (P93839) G/HBF-1, partial (72%) 52 2e-07
TC234680 similar to UP|Q9AT29 (Q9AT29) BZip transcription factor... 52 3e-07
TC216016 similar to UP|P93839 (P93839) G/HBF-1, partial (61%) 51 5e-07
TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor... 50 9e-07
BI970547 weakly similar to GP|29027731|dbj bZIP transcription fa... 49 2e-06
TC218403 UP|Q39895 (Q39895) TGACG-motif binding factor, complete 49 2e-06
TC208132 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription fact... 49 3e-06
TC232589 similar to UP|Q8LK78 (Q8LK78) ABA response element bind... 48 5e-06
CD412386 47 6e-06
>TC208968 similar to UP|GBF4_ARATH (P42777) G-box binding factor 4
(AtbZIP40), partial (27%)
Length = 1199
Score = 135 bits (340), Expect = 2e-32
Identities = 97/250 (38%), Positives = 134/250 (52%), Gaps = 9/250 (3%)
Frame = +2
Query: 1 MASSKMMMSSSKSLDHQQPSSPPPSSMPQFTTTNFID--HADSTLTDPYVPRTVDDVWQE 58
MASSK++ ++ + SS S+ P ++ + H + P++VDDVW +
Sbjct: 68 MASSKVVSTNPDLPRNSSSSSSSSSTSPISPLSSLLSDLHPMDDILKSISPKSVDDVWND 247
Query: 59 IIAGDH-----HQCKEEIPDEMMTLEDFLVKAGAVDGDDEDDEVKMSMPLTERLSGVFSL 113
I+ G + D MTLEDFL KA +++V+ + P
Sbjct: 248 IVTGATVHDAVSATTTDNADAAMTLEDFLTKA------IREEDVRGAPPPPP------PP 391
Query: 114 DGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRG--RPVVENLDKAAQQRQRRMIKNR 171
SSF +G S+SV F NGV ++G R V E +DKA Q+QRRMIKNR
Sbjct: 392 PPSSFLPFPA-DGSSSSVEPFANGVSAAPSNSVQKGKRRAVEEPVDKATLQKQRRMIKNR 568
Query: 172 ESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPP 231
ESAARSRERKQAY ELE L +LE+EN +LL E+AE ++R KQL E +IP+ +P
Sbjct: 569 ESAARSRERKQAYTSELEYLVHQLEQENVQLLNEEAEMRRQRKKQLFECIIPIEVMPKPK 748
Query: 232 RPLRRINSMQ 241
+ LRR+NS Q
Sbjct: 749 KKLRRVNSAQ 778
>TC222296 similar to UP|P93426 (P93426) Leucine zipper protein, partial (24%)
Length = 945
Score = 94.4 bits (233), Expect = 4e-20
Identities = 81/209 (38%), Positives = 103/209 (48%), Gaps = 3/209 (1%)
Frame = +3
Query: 1 MASSKMMMSSSKSLDHQQPSSPPPSSMPQFTTTNFIDHADSTLTDPYVPRTVDDVWQEII 60
MASSK++ ++ SS SS+ ++ H L P +VDD W+ I
Sbjct: 66 MASSKLVSTNPDLPRDPSSSSSSSSSISPLSSLLSDLHPMDDLLKSITPNSVDDFWKGIA 245
Query: 61 AGDHHQCKEEIPDEMMTLEDFLVKAGAVDGDDEDDEVKMSMPLTERLSGVFSLDGSSFQG 120
A +TLEDFL KA V +D V+ + P SF
Sbjct: 246 AASTDNAGG------VTLEDFLTKAIPVTEED----VRGAPP-----------PPPSFLP 362
Query: 121 MEGVEGGSASVGGFGN-GVEVVEGTRGKRG--RPVVENLDKAAQQRQRRMIKNRESAARS 177
EG S+SV F N GV ++G R V E +DKA Q+ RRMIKNRESAARS
Sbjct: 363 FPA-EGSSSSVEPFANNGVGSAPSNSVQKGKRRAVEEPVDKATLQKLRRMIKNRESAARS 539
Query: 178 RERKQAYQVELESLAVKLEEENDKLLKEK 206
RERKQAY ELE L +LE+EN +LLKE+
Sbjct: 540 RERKQAYTSELEYLVHQLEQENARLLKEE 626
Score = 26.9 bits (58), Expect = 8.3
Identities = 22/67 (32%), Positives = 31/67 (45%), Gaps = 3/67 (4%)
Frame = -2
Query: 108 SGVFSLDGSSFQGMEGVEGGSASVGGFGNG---VEVVEGTRGKRGRPVVENLDKAAQQRQ 164
S VF DG G E +EG +A + G G EVV+G G VV + ++R+
Sbjct: 329 SDVFLCDGDGL-GEEILEGHAAGIVGGGGSDPLPEVVDGIGGDGLEEVVHGVQIGEKRRK 153
Query: 165 RRMIKNR 171
R + R
Sbjct: 152 GRYRRRR 132
>TC229348 homologue to UP|Q94KA6 (Q94KA6) BZIP transcription factor 6,
partial (34%)
Length = 786
Score = 78.2 bits (191), Expect = 3e-15
Identities = 47/97 (48%), Positives = 64/97 (65%)
Frame = +1
Query: 146 GKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKE 205
G RGR ++K ++RQRRMIKNRESAARSR RKQAY +ELE+ KL+EEN++L K+
Sbjct: 196 GLRGRKSGGAVEKVIERRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENEELQKK 375
Query: 206 KAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
+AE + + Q+ E + E +R R LRR + W
Sbjct: 376 QAEIMEIQKNQVKEMMNLQREVKR--RRLRRTQTGPW 480
>TC232349 similar to UP|O81241 (O81241) Dc3 promoter-binding factor-3
(Fragment), partial (50%)
Length = 834
Score = 70.9 bits (172), Expect = 5e-13
Identities = 41/96 (42%), Positives = 57/96 (58%)
Frame = +3
Query: 144 TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLL 203
T G++ + ++K ++RQ+RMIKNRESAARSR RKQAY ELE+ +LEEEN++L
Sbjct: 357 TPGRKKSTSEDMIEKTVERRQKRMIKNRESAARSRARKQAYTNELENKVSRLEEENERLR 536
Query: 204 KEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
K K +E+++ P LRRI S
Sbjct: 537 KRKE----------LEQMLSCAPPPEPKYQLRRIAS 614
>TC222446 similar to UP|O81241 (O81241) Dc3 promoter-binding factor-3
(Fragment), partial (46%)
Length = 765
Score = 68.9 bits (167), Expect = 2e-12
Identities = 69/203 (33%), Positives = 90/203 (43%), Gaps = 39/203 (19%)
Frame = +3
Query: 76 MTLEDFLVKAGAVDGDDEDDEVKMSMPLTE--RLSGVFSLDGSSFQG------------- 120
M LEDFLVKAG + ++P T+ +SGV S SS G
Sbjct: 27 MXLEDFLVKAGVI---------AEALPTTKDRAMSGVDSNGASSQHGHWLQYQQLPSSVQ 179
Query: 121 ----MEGVEGGSASVGGFGNGVEVV------------------EGTRGKRGRPVVENLDK 158
M G G A F GV +V T G++ ++K
Sbjct: 180 QPNVMGGYVAGHAIQQPFQVGVNLVLDAAYSETPASLKGALSDTQTLGRKRGVSGIVVEK 359
Query: 159 AAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLM 218
++RQ+RMIKNRESAARSR R+QAY ELE +LEEEN++L R E M
Sbjct: 360 TVERRQKRMIKNRESAARSRARRQAYTQELEIKVSRLEEENERL-----RRLNE-----M 509
Query: 219 EEVIPVV--EKRRPPRPLRRINS 239
E +P V + +P LRR +S
Sbjct: 510 ERALPSVPPPEPKPKHQLRRTSS 578
>TC213541 similar to UP|Q9LES3 (Q9LES3) Promoter-binding factor-like protein
(ABA-responsive element binding protein 3) (AREB3),
partial (31%)
Length = 741
Score = 65.5 bits (158), Expect = 2e-11
Identities = 39/96 (40%), Positives = 54/96 (55%)
Frame = +1
Query: 144 TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLL 203
T G++ ++K ++RQ+RMIKNRESAARSR RKQAY ELE +LEEEN++L
Sbjct: 10 TAGRKRDASGNVVEKIVERRQKRMIKNRESAARSRARKQAYTQELEIKVSQLEEENERLR 189
Query: 204 KEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINS 239
++ +E +P P LRR +S
Sbjct: 190 RQNE----------IERALPSAPPPDPKHQLRRTSS 267
>BQ610152
Length = 431
Score = 63.9 bits (154), Expect = 6e-11
Identities = 31/52 (59%), Positives = 39/52 (74%)
Frame = +2
Query: 191 LAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
L LEEEN LLK++A+R ++RF QLME +IPV EKR+P LRR+NS QW
Sbjct: 2 LVTHLEEENAILLKQEADRKRQRFNQLMECLIPVEEKRKPKPMLRRVNSSQW 157
>BG155969 similar to PIR|T12622|T126 Dc3 promoter-binding factor-2 - common
sunflower, partial (43%)
Length = 414
Score = 63.9 bits (154), Expect = 6e-11
Identities = 40/85 (47%), Positives = 53/85 (62%), Gaps = 2/85 (2%)
Frame = +2
Query: 157 DKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQ 216
+K ++RQ+RMIKNRESAARSR R+QAY ELE +LEEEN++L R E
Sbjct: 2 EKTVERRQKRMIKNRESAARSRARRQAYTQELEIKVSRLEEENERL-----RRLNE---- 154
Query: 217 LMEEVIPVV--EKRRPPRPLRRINS 239
ME +P V + +P + LRR +S
Sbjct: 155 -MERALPSVPPPEPKPKQQLRRTSS 226
>CO983201
Length = 774
Score = 61.2 bits (147), Expect = 4e-10
Identities = 35/63 (55%), Positives = 40/63 (62%)
Frame = -3
Query: 144 TRGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLL 203
TR R R + +K ++R RR IKNRE AARSR RKQAY EL +LEEEN KL
Sbjct: 769 TRLGRKRDXXDAYEKTLERRLRRKIKNREXAARSRARKQAYHNELVGKVSRLEEENVKLK 590
Query: 204 KEK 206
KEK
Sbjct: 589 KEK 581
>NP004994 G/HBF-1
Length = 1137
Score = 52.8 bits (125), Expect = 1e-07
Identities = 27/51 (52%), Positives = 34/51 (65%)
Frame = +1
Query: 154 ENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLK 204
+ +D A +R RRM+ NRESA RSR RKQA+ ELE+ +L EN LLK
Sbjct: 499 QGMDPADAKRVRRMLSNRESARRSRRRKQAHLTELETQVSQLRVENSSLLK 651
>TC232503 UP|Q39896 (Q39896) TGACG-motif-binding factor, partial (97%)
Length = 1263
Score = 52.4 bits (124), Expect = 2e-07
Identities = 39/133 (29%), Positives = 69/133 (51%)
Frame = +3
Query: 90 GDDEDDEVKMSMPLTERLSGVFSLDGSSFQGMEGVEGGSASVGGFGNGVEVVEGTRGKRG 149
G + D+E++ + +G S+ Q G G+ V G G G + KRG
Sbjct: 618 GPESDEEIRRVPEIGGESAGT-----SASQPDAGSNAGTERVQGTGEGQK-------KRG 761
Query: 150 RPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAER 209
R + K R +R+++NR SA ++RERK+AY ++LE+ LE++N + LKE+
Sbjct: 762 RSPADKESK----RLKRLLRNRVSAQQARERKKAYLIDLETRVKDLEKKNSE-LKERLS- 923
Query: 210 TKERFKQLMEEVI 222
T + Q++ +++
Sbjct: 924 TLQNENQMLRQIL 962
>TC227230 similar to UP|P93839 (P93839) G/HBF-1, partial (72%)
Length = 1250
Score = 52.0 bits (123), Expect = 2e-07
Identities = 27/51 (52%), Positives = 34/51 (65%)
Frame = +2
Query: 154 ENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLK 204
+N D A +R RRM+ NRESA RSR RKQA+ +LE+ +L EN LLK
Sbjct: 479 DNTDPADVKRVRRMLSNRESARRSRRRKQAHLTDLETQVSQLRGENSTLLK 631
>TC234680 similar to UP|Q9AT29 (Q9AT29) BZip transcription factor, partial
(49%)
Length = 571
Score = 51.6 bits (122), Expect = 3e-07
Identities = 33/83 (39%), Positives = 46/83 (54%)
Frame = +2
Query: 154 ENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAERTKER 213
+NL +++ RRMI NRESA RSR RKQ + EL S V L EN +L+ +K E
Sbjct: 323 QNLSLINERKHRRMISNRESARRSRMRKQKHLDELWSQVVWLRNENHQLM-DKLNHVSES 499
Query: 214 FKQLMEEVIPVVEKRRPPRPLRR 236
Q+M+E + E+ R + R
Sbjct: 500 HDQVMQENAQLKEQALELRQMIR 568
>TC216016 similar to UP|P93839 (P93839) G/HBF-1, partial (61%)
Length = 1764
Score = 50.8 bits (120), Expect = 5e-07
Identities = 26/50 (52%), Positives = 32/50 (64%)
Frame = +3
Query: 155 NLDKAAQQRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLK 204
N+ +R RRM+ NRESA RSR RKQA+ ELE+ +L EN LLK
Sbjct: 714 NMTPVDAKRVRRMLSNRESARRSRRRKQAHLTELETQVSQLRSENSSLLK 863
>TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor, partial
(17%)
Length = 792
Score = 50.1 bits (118), Expect = 9e-07
Identities = 26/47 (55%), Positives = 32/47 (67%)
Frame = +3
Query: 162 QRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAE 208
QR R+IKNRESA RSR RKQAY+ LE +L EEN +L ++ E
Sbjct: 336 QRHARIIKNRESAVRSRARKQAYRKGLEVEIARLTEENSRLKRQLKE 476
>BI970547 weakly similar to GP|29027731|dbj bZIP transcription factor
{Arabidopsis thaliana}, partial (15%)
Length = 697
Score = 49.3 bits (116), Expect = 2e-06
Identities = 26/47 (55%), Positives = 32/47 (67%)
Frame = -1
Query: 162 QRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKLLKEKAE 208
QR R+IKNRESA RSR RKQAY+ LE +L EEN +L ++ E
Sbjct: 490 QRHTRVIKNRESAVRSRARKQAYRKGLEVEISRLTEENSRLKRQLKE 350
>TC218403 UP|Q39895 (Q39895) TGACG-motif binding factor, complete
Length = 1195
Score = 49.3 bits (116), Expect = 2e-06
Identities = 32/93 (34%), Positives = 55/93 (58%), Gaps = 3/93 (3%)
Frame = +1
Query: 133 GFGNGVEVVEGT---RGKRGRPVVENLDKAAQQRQRRMIKNRESAARSRERKQAYQVELE 189
G G E +GT + KRGR + K R +R+++NR SA ++RERK+AY ++LE
Sbjct: 709 GSNAGTERAQGTGDSQKKRGRSPADKESK----RLKRLLRNRVSAQQARERKKAYLIDLE 876
Query: 190 SLAVKLEEENDKLLKEKAERTKERFKQLMEEVI 222
+ LE++N + LKE+ T + Q++ +++
Sbjct: 877 TRVKDLEKKNSE-LKERLS-TLQNENQMLRQIL 969
>TC208132 homologue to UP|Q94KA7 (Q94KA7) BZIP transcription factor 3,
partial (33%)
Length = 706
Score = 48.5 bits (114), Expect = 3e-06
Identities = 40/111 (36%), Positives = 53/111 (47%), Gaps = 10/111 (9%)
Frame = +1
Query: 127 GSASVGGFGNGVEVVEGTRGKRGRPVVENLDKAAQQ----------RQRRMIKNRESAAR 176
GS+++ G G V T G V + D A Q RQRR NRESA R
Sbjct: 58 GSSNIPGLGRKVP---STAVAGGMVTVGSRDSAQSQLWLQDERELKRQRRKQSNRESARR 228
Query: 177 SRERKQAYQVELESLAVKLEEENDKLLKEKAERTKERFKQLMEEVIPVVEK 227
SR RKQA EL A L+EEN L+ + R + ++QL+ E + E+
Sbjct: 229 SRLRKQAECDELAQRAEALKEENAS-LRSEVNRIRSDYEQLLSENAALKER 378
>TC232589 similar to UP|Q8LK78 (Q8LK78) ABA response element binding factor,
partial (10%)
Length = 464
Score = 47.8 bits (112), Expect = 5e-06
Identities = 25/41 (60%), Positives = 29/41 (69%)
Frame = +2
Query: 162 QRQRRMIKNRESAARSRERKQAYQVELESLAVKLEEENDKL 202
QR R+IKNRESA RSR RKQAY+ LE +L EEN +L
Sbjct: 341 QRHTRVIKNRESAVRSRARKQAYRKGLEVEISRLTEENSRL 463
>CD412386
Length = 576
Score = 47.4 bits (111), Expect = 6e-06
Identities = 20/35 (57%), Positives = 28/35 (79%)
Frame = -2
Query: 208 ERTKERFKQLMEEVIPVVEKRRPPRPLRRINSMQW 242
ER KER+KQLME+V+P+ +K++P LRR S+QW
Sbjct: 575 ERKKERYKQLMEKVLPIAQKQKPLCILRRARSLQW 471
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,409,111
Number of Sequences: 63676
Number of extensions: 102423
Number of successful extensions: 1186
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 1130
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1178
length of query: 242
length of database: 12,639,632
effective HSP length: 95
effective length of query: 147
effective length of database: 6,590,412
effective search space: 968790564
effective search space used: 968790564
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0052a.3


