

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0048.13
(98 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
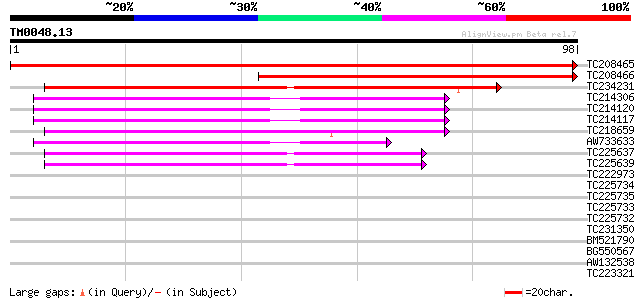
Score E
Sequences producing significant alignments: (bits) Value
TC208465 similar to GB|AAM16215.1|20334918|AY094059 At1g65700/F1... 170 1e-43
TC208466 similar to GB|AAM16215.1|20334918|AY094059 At1g65700/F1... 94 8e-21
TC234231 weakly similar to UP|Q9W2K2 (Q9W2K2) CG4279-PA, partial... 62 5e-11
TC214306 homologue to UP|RUXG_ARATH (O82221) Probable small nucl... 47 1e-06
TC214120 homologue to UP|RUXG_ARATH (O82221) Probable small nucl... 47 2e-06
TC214117 homologue to UP|RUXG_ARATH (O82221) Probable small nucl... 47 2e-06
TC218659 weakly similar to UP|Q9VJI7 (Q9VJI7) CG13277-PA (RH7352... 46 3e-06
AW733633 43 3e-05
TC225637 homologue to UP|Q9FKB0 (Q9FKB0) Sm-like protein, partia... 40 2e-04
TC225639 homologue to UP|Q9FKB0 (Q9FKB0) Sm-like protein, partia... 40 2e-04
TC222973 similar to UP|Q945P8 (Q945P8) At1g19120/F14D16_26, part... 35 0.005
TC225734 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucle... 33 0.017
TC225735 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucle... 33 0.017
TC225733 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucle... 33 0.023
TC225732 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucle... 33 0.023
TC231350 homologue to PIR|T10586|T10586 small nuclear ribonucleo... 31 0.086
BM521790 homologue to PIR|T10586|T10 small nuclear ribonucleopro... 31 0.086
BG550567 similar to GP|21593343|gb|A small nuclear ribonucleopro... 31 0.11
AW132538 homologue to GP|9758886|dbj|B U6 snRNA-associated Sm-li... 31 0.11
TC223321 similar to GB|CAD41024.1|21741213|OSJN00063 OSJNBb0086G... 30 0.15
>TC208465 similar to GB|AAM16215.1|20334918|AY094059 At1g65700/F1E22_3
{Arabidopsis thaliana;} , complete
Length = 684
Score = 170 bits (430), Expect = 1e-43
Identities = 85/98 (86%), Positives = 90/98 (91%)
Frame = +3
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALG 60
MS GPGLESLVDQ ISVITNDGRNIVG LKGFDQATNIILDESHERV+STKEGVQ + LG
Sbjct: 120 MSAGPGLESLVDQTISVITNDGRNIVGVLKGFDQATNIILDESHERVYSTKEGVQQLVLG 299
Query: 61 LYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKPVIH 98
LYIIRGDNISVVGELDE+LD +LD SK+RA PLKPVIH
Sbjct: 300 LYIIRGDNISVVGELDEELDSSLDLSKLRAHPLKPVIH 413
>TC208466 similar to GB|AAM16215.1|20334918|AY094059 At1g65700/F1E22_3
{Arabidopsis thaliana;} , partial (56%)
Length = 435
Score = 94.4 bits (233), Expect = 8e-21
Identities = 45/55 (81%), Positives = 50/55 (90%)
Frame = +1
Query: 44 HERVFSTKEGVQLIALGLYIIRGDNISVVGELDEDLDKNLDKSKIRAQPLKPVIH 98
HERV+STKEGVQ + LGLYIIRGDNISVVGELDE+LD +LD SK+RA PLKPVIH
Sbjct: 13 HERVYSTKEGVQQLVLGLYIIRGDNISVVGELDEELDSSLDLSKLRAHPLKPVIH 177
>TC234231 weakly similar to UP|Q9W2K2 (Q9W2K2) CG4279-PA, partial (50%)
Length = 748
Score = 62.0 bits (149), Expect = 5e-11
Identities = 33/82 (40%), Positives = 55/82 (66%), Gaps = 3/82 (3%)
Frame = +2
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L S +D+++ V+ DGR ++GTL+ FDQ N +L+ + ERV + I LGLY+IRG
Sbjct: 197 LASYLDKKLLVLLRDGRKLMGTLRSFDQFANAVLEGACERVI-VGDLYCDIPLGLYVIRG 373
Query: 67 DNISVVGELD---EDLDKNLDK 85
+N+ ++GELD E+L +++ +
Sbjct: 374 ENVVLIGELDLEREELPEHMTR 439
>TC214306 homologue to UP|RUXG_ARATH (O82221) Probable small nuclear
ribonucleoprotein G (snRNP-G) (Sm protein G) (Sm-G)
(SmG), partial (98%)
Length = 797
Score = 47.4 bits (111), Expect = 1e-06
Identities = 24/72 (33%), Positives = 43/72 (59%)
Frame = +3
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+++ + N R IVGTL+GFDQ N+++D + E G + +G+ +I
Sbjct: 300 PDLKKYMDKKLQIKLNGNRMIVGTLRGFDQFMNLVVDNTVE-----VNGNEKNDIGMVVI 464
Query: 65 RGDNISVVGELD 76
RG+++ V L+
Sbjct: 465 RGNSVVTVEALE 500
>TC214120 homologue to UP|RUXG_ARATH (O82221) Probable small nuclear
ribonucleoprotein G (snRNP-G) (Sm protein G) (Sm-G)
(SmG), partial (98%)
Length = 583
Score = 46.6 bits (109), Expect = 2e-06
Identities = 23/72 (31%), Positives = 43/72 (58%)
Frame = +2
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+++ + N R +VGTL+GFDQ N+++D + E G + +G+ +I
Sbjct: 110 PDLKKYMDKKLQIKLNANRMVVGTLRGFDQFMNLVVDNTVE-----VNGNEKNDIGMVVI 274
Query: 65 RGDNISVVGELD 76
RG+++ V L+
Sbjct: 275 RGNSVVTVEALE 310
>TC214117 homologue to UP|RUXG_ARATH (O82221) Probable small nuclear
ribonucleoprotein G (snRNP-G) (Sm protein G) (Sm-G)
(SmG), partial (98%)
Length = 534
Score = 46.6 bits (109), Expect = 2e-06
Identities = 23/72 (31%), Positives = 43/72 (58%)
Frame = +1
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+++ + N R +VGTL+GFDQ N+++D + E G + +G+ +I
Sbjct: 106 PDLKKYMDKKLQIKLNANRMVVGTLRGFDQFMNLVVDNTVE-----VNGNEKNDIGMVVI 270
Query: 65 RGDNISVVGELD 76
RG+++ V L+
Sbjct: 271 RGNSVVTVEALE 306
>TC218659 weakly similar to UP|Q9VJI7 (Q9VJI7) CG13277-PA (RH73529p), partial
(81%)
Length = 578
Score = 45.8 bits (107), Expect = 3e-06
Identities = 26/74 (35%), Positives = 39/74 (52%), Gaps = 4/74 (5%)
Frame = +3
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGV----QLIALGLY 62
L VD+ + V GR + GTLKG+DQ N++LDE+ E + + + Q LGL
Sbjct: 144 LAKFVDKGVQVKLTGGRQVTGTLKGYDQLLNLVLDEAVEFLRDPDDPLKTTDQTRNLGLI 323
Query: 63 IIRGDNISVVGELD 76
+ RG + +V D
Sbjct: 324 VCRGTAVMLVSPTD 365
>AW733633
Length = 289
Score = 42.7 bits (99), Expect = 3e-05
Identities = 21/62 (33%), Positives = 37/62 (58%)
Frame = +3
Query: 5 PGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
P L+ +D+++ + N R +VGTL+GFDQ N+++D + E G + +G+ +I
Sbjct: 60 PDLKKYMDKKLQIKLNANRMVVGTLRGFDQFMNLVVDNTVE-----VNGNEKNDIGMVVI 224
Query: 65 RG 66
RG
Sbjct: 225 RG 230
>TC225637 homologue to UP|Q9FKB0 (Q9FKB0) Sm-like protein, partial (97%)
Length = 591
Score = 40.0 bits (92), Expect = 2e-04
Identities = 20/66 (30%), Positives = 39/66 (58%)
Frame = +3
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
++ + +I VI + +VGTL+GFD N++L++ E T EG ++ L ++ G
Sbjct: 180 IDRCIGSKIWVIMKGDKELVGTLRGFDVYANMVLEDVTEYEI-TAEGRRITKLDQILLNG 356
Query: 67 DNISVV 72
+NI+++
Sbjct: 357 NNIAIL 374
>TC225639 homologue to UP|Q9FKB0 (Q9FKB0) Sm-like protein, partial (97%)
Length = 830
Score = 40.0 bits (92), Expect = 2e-04
Identities = 20/66 (30%), Positives = 39/66 (58%)
Frame = +1
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
++ + +I VI + +VGTL+GFD N++L++ E T EG ++ L ++ G
Sbjct: 361 IDRCIGSKIWVIMKGDKELVGTLRGFDVYVNMVLEDVTEYEI-TAEGRRITKLDQILLNG 537
Query: 67 DNISVV 72
+NI+++
Sbjct: 538 NNIAIL 555
>TC222973 similar to UP|Q945P8 (Q945P8) At1g19120/F14D16_26, partial (37%)
Length = 457
Score = 35.4 bits (80), Expect = 0.005
Identities = 14/33 (42%), Positives = 23/33 (69%)
Frame = +1
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNII 39
L S +D+++ V+ DGR ++GTL+ FDQ N +
Sbjct: 358 LASYLDKKLLVLLRDGRKLMGTLRSFDQFANAV 456
>TC225734 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucleoprotein
homolog, complete
Length = 837
Score = 33.5 bits (75), Expect = 0.017
Identities = 20/66 (30%), Positives = 34/66 (51%)
Frame = +2
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L+S QI + I G + GFD+ N++LD++ E K LG +++G
Sbjct: 167 LQSKARIQIWLFEQKDLRIEGRIIGFDEYMNLVLDDAEEVNIKKKSRK---TLGRILLKG 337
Query: 67 DNISVV 72
DNI+++
Sbjct: 338 DNITLM 355
>TC225735 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucleoprotein
homolog, complete
Length = 431
Score = 33.5 bits (75), Expect = 0.017
Identities = 20/66 (30%), Positives = 34/66 (51%)
Frame = +1
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L+S QI + I G + GFD+ N++LD++ E K LG +++G
Sbjct: 205 LQSKARIQIWLFEQKDLRIEGRIIGFDEYMNLVLDDAEEVNIKKKSRK---TLGKILLKG 375
Query: 67 DNISVV 72
DNI+++
Sbjct: 376 DNITLM 393
>TC225733 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucleoprotein
homolog, complete
Length = 530
Score = 33.1 bits (74), Expect = 0.023
Identities = 20/66 (30%), Positives = 34/66 (51%)
Frame = +2
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L+S QI + I G + GFD+ N++LD++ E K LG +++G
Sbjct: 140 LQSKARIQIWLFEQKDLRIEGRIIGFDEYMNLVLDDAEEVNVKKKSRK---TLGRILLKG 310
Query: 67 DNISVV 72
DNI+++
Sbjct: 311 DNITLM 328
>TC225732 homologue to UP|Q8LAK5 (Q8LAK5) Small nuclear ribonucleoprotein
homolog, complete
Length = 715
Score = 33.1 bits (74), Expect = 0.023
Identities = 20/66 (30%), Positives = 34/66 (51%)
Frame = +1
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRG 66
L+S QI + I G + GFD+ N++LD++ E K LG +++G
Sbjct: 160 LQSKARIQIWLFEQKDLRIEGRIIGFDEYMNLVLDDAEEVNVKKKSRK---TLGRILLKG 330
Query: 67 DNISVV 72
DNI+++
Sbjct: 331 DNITLM 348
>TC231350 homologue to PIR|T10586|T10586 small nuclear
ribonucleoprotein-associated protein homolog F9F13.90 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(60%)
Length = 662
Score = 31.2 bits (69), Expect = 0.086
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 10/79 (12%)
Frame = +2
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHE-RVFSTKEGVQLI-- 57
MS + ++ ++ V DGR +VG FD+ N++L + E R +G +
Sbjct: 104 MSKSSKMLQYINYRMRVTIQDGRQLVGKFMAFDRHMNLVLGDCEEFRKLPPAKGKKPAEG 283
Query: 58 -------ALGLYIIRGDNI 69
LGL ++RG+ +
Sbjct: 284 ADREDRRTLGLVLLRGEEV 340
>BM521790 homologue to PIR|T10586|T10 small nuclear
ribonucleoprotein-associated protein homolog F9F13.90 -
Arabidopsis thaliana, partial (46%)
Length = 428
Score = 31.2 bits (69), Expect = 0.086
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 10/79 (12%)
Frame = +3
Query: 1 MSTGPGLESLVDQQISVITNDGRNIVGTLKGFDQATNIILDESHE-RVFSTKEGVQLI-- 57
MS + ++ ++ V DGR +VG FD+ N++L + E R +G +
Sbjct: 60 MSKSSKMLQYINYRMRVTIQDGRQLVGKFMAFDRHMNLVLGDCEEFRKLPPAKGKKPAEG 239
Query: 58 -------ALGLYIIRGDNI 69
LGL ++RG+ +
Sbjct: 240 GDREDRRTLGLVLLRGEEV 296
>BG550567 similar to GP|21593343|gb|A small nuclear ribonucleoprotein homolog
{Arabidopsis thaliana}, partial (82%)
Length = 379
Score = 30.8 bits (68), Expect = 0.11
Identities = 17/48 (35%), Positives = 31/48 (64%)
Frame = +3
Query: 25 IVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYIIRGDNISVV 72
I G + GFD+ N++LD+ E+V K+ + LG +++GDNI+++
Sbjct: 195 IEGRIIGFDEYMNLVLDDK-EKVNIKKKSRK--TLGRILLKGDNITLM 329
>AW132538 homologue to GP|9758886|dbj|B U6 snRNA-associated Sm-like
protein-like {Arabidopsis thaliana}, partial (57%)
Length = 429
Score = 30.8 bits (68), Expect = 0.11
Identities = 12/36 (33%), Positives = 23/36 (63%)
Frame = +2
Query: 7 LESLVDQQISVITNDGRNIVGTLKGFDQATNIILDE 42
++ + +I VI + +VGTL+GFD N++L++
Sbjct: 314 IDRCIGSKIWVIMKGDKELVGTLRGFDVYVNMVLED 421
>TC223321 similar to GB|CAD41024.1|21741213|OSJN00063 OSJNBb0086G13.8 {Oryza
sativa (japonica cultivar-group);} , partial (15%)
Length = 724
Score = 30.4 bits (67), Expect = 0.15
Identities = 20/74 (27%), Positives = 41/74 (55%), Gaps = 1/74 (1%)
Frame = +1
Query: 6 GLESLVDQ-QISVITNDGRNIVGTLKGFDQATNIILDESHERVFSTKEGVQLIALGLYII 64
G+E V++ I+ I +DG ++VG KG+ Q +L + + + T V++ ++G + +
Sbjct: 82 GMEGKVERIWITDIASDGVHLVGHTKGYIQ----VLVLAPDNMLGTSAMVKITSVGRWSV 249
Query: 65 RGDNISVVGELDED 78
GD I V + ++
Sbjct: 250 FGDVIETVNPVSDN 291
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.138 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,687,226
Number of Sequences: 63676
Number of extensions: 22659
Number of successful extensions: 95
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 94
length of query: 98
length of database: 12,639,632
effective HSP length: 74
effective length of query: 24
effective length of database: 7,927,608
effective search space: 190262592
effective search space used: 190262592
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0048.13


