

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0045.8
(1555 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
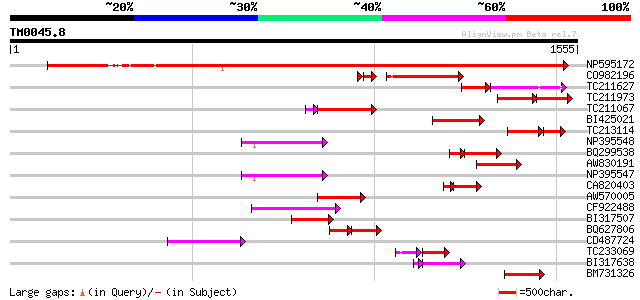
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 1149 0.0
CO982196 249 2e-73
TC211627 125 5e-52
TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, parti... 120 8e-48
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 154 1e-40
BI425021 163 6e-40
TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, parti... 120 8e-38
NP395548 reverse transcriptase [Glycine max] 155 2e-37
BQ299538 108 3e-36
AW830191 148 2e-35
NP395547 reverse transcriptase [Glycine max] 148 2e-35
CA820403 weakly similar to GP|13273463|gb| pol protein integrase... 109 5e-33
AW570005 139 1e-32
CF922488 133 7e-31
BI317507 125 1e-28
BQ627806 73 1e-26
CD487724 112 1e-24
TC233069 84 6e-23
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 98 8e-23
BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza... 102 1e-21
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 1149 bits (2971), Expect = 0.0
Identities = 624/1444 (43%), Positives = 878/1444 (60%), Gaps = 14/1444 (0%)
Frame = +1
Query: 103 RVDIPMFNGNDAYGWITKVERFFRLSRVEETEKVEMVMIAMEDRALSWFQWWEEQAPVRA 162
++D P F+G + WI K E+FF + +++ + + ++ + W+Q ++ P +
Sbjct: 295 KLDFPRFDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWYQMLQKTEPFSS 474
Query: 163 WEPFKQALIRRFQPELVQNPFGPLLSAKQKGSVMEYREHFELLAAPMRNADREVLKGVFL 222
W+ F +AL F P P L Q +V EY F L + E + F+
Sbjct: 475 WQAFTRALELDFGPSAYDCPRATLFKLNQSATVNEYYMQFTALVNRVDGLSAEAILDCFV 654
Query: 223 NGLHEEIKAEMKLYPSEDLADLMDRALLLEEKNSAMRGGKSKEEDKRGWKEKGGFGGRFS 282
+GL EEI ++K L + A L EEK ++ K+ R F+
Sbjct: 655 SGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPKTKTFSNLARN----------FT 804
Query: 283 TNSGETRGRSIYPNVSYQSKGAGGTQGNEGKPQHLKEGTENTNNEGKIPERKWNGG-QRL 341
+N+ T+ YP + Q N+ +L + + P N +++
Sbjct: 805 SNTSATQK---YPPTN---------QKNDNPKPNLPPLLPTPSTK---PFNLRNQNIKKI 939
Query: 342 TQTELQERSRRGLCFKCGEKWGREHICAKKNFQLILIEGEDEEEEEEVFEEAEDGEFVLE 401
+ E+Q R + LC+ C EK+ H C + L+ +E DE++ +E E+ ++
Sbjct: 940 SPAEIQLRREKNLCYFCDEKFSPAHKCPNRQVMLLQLEETDEDQTDEQVMVTEEAN--MD 1113
Query: 402 GKVLQLSLNSKEGLTSNRSFKVKGKIGEREILILVDCGATSNFISQELVAELEIPVVATS 461
LSLN+ G + + G++G + ILVD G++ NFI + L++PV
Sbjct: 1114 DDTHHLSLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAP 1293
Query: 462 EYVVEVGNGARERNSGVCKNLKLEVQGIPIIQHFFILGLGGTELVLGMDWLASLGNIEAN 521
V VGNG G+ + L L +QG + ++L + G +++LG WLA+LG A+
Sbjct: 1294 NLRVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVAD 1473
Query: 522 FQDLIIKWELNGQKMCMQGEPSFCKVAATWKSIKKTKHDEG-EEYFLSYECSEEEPTANV 580
+ L +K+ N + + +QGE + A ++ ++ + EE F +E P +
Sbjct: 1474 YAALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKEVPEDTL 1653
Query: 581 TI------PELWIKLLTEFPEVFQEPKELPPKRATDHAILLQEGAPIPNIRPYRYPFYQK 634
PEL I LL + +VF P LPP+R DHAI L++G+ +RPYRYP QK
Sbjct: 1654 KDLPTNIDPELAI-LLHTYAQVFAVPASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQK 1830
Query: 635 NEIEKLVKEMLAAGIIRHSTSPFSSPAILVKKKDGGWRFCVDYRALNKVTIPDKFPIPII 694
++IEK+++EML GII+ S SPFS P +LVKKKDG WRFC DYRALN +T+ D FP+P +
Sbjct: 1831 DQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTV 2010
Query: 695 DELLDEIGTAEIFSKLDLKSGYHQIRMREEDIPKTAFRTHEGHYEYLVLPFGLTNAPSTF 754
DELLDE+ A+ FSKLDL+SGYHQI ++ ED KTAFRTH GHYE+LV+PFGLTNAP+TF
Sbjct: 2011 DELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATF 2190
Query: 755 QALMNQVLRPYLRKFVLVFFYDILIYSNNVDLHKEHLREVLQVLRDNHLVANQKKCSFGQ 814
Q LMN++ + LRKFVLVFF DILIYS + H +HL VLQ L+ + L A KCSFG
Sbjct: 2191 QCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGD 2370
Query: 815 SELIYLGHVISKEGVAADPSKIKDMLNWPLPKDVKGLRGFLGLTGYYRRFVRNYSKLAQP 874
+E+ YLGH +S GV+ + +K++ +L+WP P +VK LRGFLGLTGYYRRF+++Y+ +A P
Sbjct: 2371 TEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAGP 2550
Query: 875 LNQLLKKNNFSWSAGATQAFDKLKEIMTTVPVLAVPDFQKTFVLETDASGKGLGAVLMQG 934
L LL+K++F W+ A AF KLK+ MT PVL++PDF + F+LETDASG G+GAVL Q
Sbjct: 2551 LTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQN 2730
Query: 935 GRPVAYMSKTLSERAQAKSVYERELMAVVLAVQKWRHYLLGCKFIVHTDQKSLRFLAEQR 994
G P+AY SK L+ R Q +S Y REL+A+ A+ K+RHYLLG KFI+ TDQ+SL+ L +Q
Sbjct: 2731 GHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQS 2910
Query: 995 LMGEEQQKWVSKLMGFDFEIKYKPGIENKAADALSRKLQFSAISSVQCEDWEDLETEILA 1054
L EQQ W+ K +G+DF+I+YKPG +N+AADALSR A S E+L +++
Sbjct: 2911 LQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSRMFML-AWSEPHSIFLEELRARLIS 3087
Query: 1055 DDKYQKIIQEITTQGPVPAGYHMRRGRLLYKNRIVLPKTSGKIPIILQEFHDSAVGGHAG 1114
D + K + E QG + Y +R G L +K+R+V+P + ILQE+H S +GGHAG
Sbjct: 3088 -DPHLKQLMETYKQGADASHYTVREGLLYWKDRVVIPAEEEIVNKILQEYHSSPIGGHAG 3264
Query: 1115 IFRTYKRISALFFWEGMKLDIQTYVQKCEICQRNKYETLNPAGYLQPLPIPSQVWSDISM 1174
I RT R+ A F+W M+ D++ Y+QKC ICQ+ K PAG LQPLPIP QVW D++M
Sbjct: 3265 ITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQAKSNNTLPAGLLQPLPIPQQVWEDVAM 3444
Query: 1175 DFIGGLPKTMGKDTILVVVDRFTKYAHFLALSHPYNAKEVAELFIKEIVKLHGFPTSIVS 1234
DFI GLP + G I+VV+DR TKYAHF+ L YN+K VAE F+ IVKLHG P SIVS
Sbjct: 3445 DFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKADYNSKVVAEAFMSHIVKLHGIPRSIVS 3624
Query: 1235 DRDRVFLSSFWSELFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGAKPKQWPK 1294
DRDRVF S+FW LFKL GT L SSAYHPQ+DGQ+EV+N+C+E YLRC T PK W K
Sbjct: 3625 DRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDGQSEVLNKCLEMYLRCFTYEHPKGWVK 3804
Query: 1295 WLSWAEFWYNTNYHSAIKTTPFQALYGREPPVIIKGTDSLASVNEVEKMTAERNLFLDTL 1354
L WAEFWYNT YH ++ TPF+ALYGREPP + + S+ EV + +R+ L L
Sbjct: 3805 ALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLTRQACSIDDPAEVREQLTDRDALLAKL 3984
Query: 1355 KENLEKAQNRMKQQANKHRRDIQLKVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPV 1414
K NL +AQ MK+QA+K R D+ ++GD V +K+QPY+ S RKNQKLS R++GP+ V
Sbjct: 3985 KINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKLQPYRQHSAVLRKNQKLSMRYFGPFKV 4164
Query: 1415 IEKINAVAYKLQLPEGSQVHPVFHVSLLKKAPNEGVNSQ---PLPVYMTEEWELKVEPES 1471
+ KI VAYKL+LP +++HPVFHVS LK P G PLP+ +TE + ++P
Sbjct: 4165 LAKIGDVAYKLELPSAARIHPVFHVSQLK--PFNGTAQDPYLPLPLTVTEMGPV-MQPVK 4335
Query: 1472 IIDSR---AGDKGTVEVLVKWKNMPEFENSWEDYTELLEQFPEHHLEDKVILQGGRDDTD 1528
I+ SR G ++LV+W+N + E +WED ++ +P +LEDKV+ +G + T+
Sbjct: 4336 ILASRIIIRGHNQIEQILVQWENGLQDEATWEDIEDIKASYPTFNLEDKVVFKGEGNVTN 4515
Query: 1529 PLVR 1532
+ R
Sbjct: 4516 GMSR 4527
>CO982196
Length = 812
Score = 249 bits (636), Expect(3) = 2e-73
Identities = 125/212 (58%), Positives = 155/212 (72%)
Frame = +1
Query: 1033 QFSAISSVQCEDWEDLETEILADDKYQKIIQEITTQGPVPAGYHMRRGRLLYKNRIVLPK 1092
QF + DWE+ EI A + +I Q I T+ GY +R G+L +K+R+VL K
Sbjct: 184 QFQWCKRRELADWEE---EIQAYLELYEIYQGILTKTTKKPGYAIRGGKLYFKDRLVLSK 354
Query: 1093 TSGKIPIILQEFHDSAVGGHAGIFRTYKRISALFFWEGMKLDIQTYVQKCEICQRNKYET 1152
S KIP++L+E DS +GGH+G FRT+KR++ + FW+GMK + YV CEIC+RNK T
Sbjct: 355 NSTKIPLLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACEICRRNKTST 534
Query: 1153 LNPAGYLQPLPIPSQVWSDISMDFIGGLPKTMGKDTILVVVDRFTKYAHFLALSHPYNAK 1212
L+PAG L LPIP++VW+DISMDFIGGLPK GKD ILVVVDR TKYAHF ALSHPY AK
Sbjct: 535 LSPAGLL*LLPIPTKVWTDISMDFIGGLPKAQGKDNILVVVDRLTKYAHFFALSHPYTAK 714
Query: 1213 EVAELFIKEIVKLHGFPTSIVSDRDRVFLSSF 1244
EVAELFIKE+V+LHGFP SIVSD R+F+S F
Sbjct: 715 EVAELFIKELVRLHGFPASIVSDXXRLFMSLF 810
Score = 46.2 bits (108), Expect(3) = 2e-73
Identities = 21/36 (58%), Positives = 27/36 (74%)
Frame = +2
Query: 970 RHYLLGCKFIVHTDQKSLRFLAEQRLMGEEQQKWVS 1005
RHY +G KFI+ T+ +S +FL EQRLM EEQ KW +
Sbjct: 53 RHYPVGKKFIIRTN*RSSKFLNEQRLMSEEQFKWAT 160
Score = 21.9 bits (45), Expect(3) = 2e-73
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +3
Query: 954 VYERELMAVVLAV 966
+YERELM VVL V
Sbjct: 6 MYERELMDVVLPV 44
>TC211627
Length = 1034
Score = 125 bits (315), Expect(2) = 5e-52
Identities = 78/216 (36%), Positives = 121/216 (55%), Gaps = 7/216 (3%)
Frame = +3
Query: 1319 LYGREPPVIIKGTDSLASVNEVEKMTAERNLFLDTLKENLEKAQNRMKQQANKHRRDIQL 1378
+YG+ PP + + ++V V+ + TL L+K Q+ MK+ A+ HRRD+
Sbjct: 243 MYGKPPPALPLYSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTF 422
Query: 1379 KVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEKINAVAYKLQLPEGSQVHPVFH 1438
+GD VY+++ PY+ S+ + KLS RFYGPY + ++ VAY+LQLP S++HP+FH
Sbjct: 423 NIGDWVYVRL*PYRQTSI-QSTYTKLSKRFYGPYQIQARVGQVAYRLQLPPTSKIHPIFH 599
Query: 1439 VSLLK----KAPNEGVNSQPLPVYMTEEWELKVEPESIIDSRAGDKGT---VEVLVKWKN 1491
VSLLK P E + LP + T L V+P +D + + T +VLV+W N
Sbjct: 600 VSLLKVHHGPIPPELL---ALPPFSTTNHPL-VQPLQFLDWKMDESTTPPIPQVLVQWTN 767
Query: 1492 MPEFENSWEDYTELLEQFPEHHLEDKVILQGGRDDT 1527
+ + +WE +T+L + + LEDKV Q G D+
Sbjct: 768 LAPEDTTWESWTQLKDIY---DLEDKVCFQTGGIDS 866
Score = 99.4 bits (246), Expect(2) = 5e-52
Identities = 45/79 (56%), Positives = 58/79 (72%)
Frame = +2
Query: 1239 VFLSSFWSELFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGAKPKQWPKWLSW 1298
+F+S W ELF ++GTKL+FS+AYHPQTDGQTEV+NR +E YLR P+ W K+LS
Sbjct: 2 IFISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSL 181
Query: 1299 AEFWYNTNYHSAIKTTPFQ 1317
AE YNT+ HS I +PF+
Sbjct: 182 AE*CYNTSVHSGIGFSPFE 238
>TC211973 weakly similar to UP|Q84ZV5 (Q84ZV5) Polyprotein, partial (4%)
Length = 730
Score = 120 bits (302), Expect(2) = 8e-48
Identities = 62/112 (55%), Positives = 80/112 (71%), Gaps = 1/112 (0%)
Frame = +1
Query: 1337 VNEVEKMTAERNLFLDTLKENLEKAQNRMKQQANKHRRDIQLKVGDMVYLKIQPYKLKSL 1396
+ EV K+ R+ L TL+ENL K+ + M NK RRDI+ VGD V+LK+QPY+ +SL
Sbjct: 13 LEEVNKLIIARDGLLATLRENLLKS*DIM*ANTNKRRRDIEYVVGD*VFLKMQPYRRRSL 192
Query: 1397 ARRKNQKLSPRFYGPYPVIEKINAVAYKLQLPEGSQVHPVFHVSLLKKA-PN 1447
A+R N+KLSPRFY P+ V K+ +AYKL LP ++HPVFHVSLLKKA PN
Sbjct: 193 AKRINEKLSPRFYAPFQVFNKVGTIAYKLDLPSHIKIHPVFHVSLLKKASPN 348
Score = 90.1 bits (222), Expect(2) = 8e-48
Identities = 47/98 (47%), Positives = 63/98 (63%)
Frame = +3
Query: 1445 APNEGVNSQPLPVYMTEEWELKVEPESIIDSRAGDKGTVEVLVKWKNMPEFENSWEDYTE 1504
+P E SQPLP ++E+W+L+ +S++DSR G V+VL++WKN+P ENSWE +
Sbjct: 324 SPQES-QSQPLPPMLSEDWKLQTYSDSVLDSRELQPGNVKVLIQWKNLPPSENSWESVAK 500
Query: 1505 LLEQFPEHHLEDKVILQGGRDDTDPLVRPRFGKVYTRK 1542
L E F +HLEDKV L GG D +P KVYTRK
Sbjct: 501 LQEIFSIYHLEDKVSLLGGGIDKHK-HKPPIPKVYTRK 611
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 154 bits (389), Expect(2) = 1e-40
Identities = 76/163 (46%), Positives = 106/163 (64%), Gaps = 1/163 (0%)
Frame = +1
Query: 843 PLPKDVKGLRGFLGLTGYYRRFVRNYSKLAQPLNQLLKKNN-FSWSAGATQAFDKLKEIM 901
P K V +R F GL +YRRFV N+S +A PLN+L+KKN F+W QAF LKE +
Sbjct: 97 PTLKSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKL 276
Query: 902 TTVPVLAVPDFQKTFVLETDASGKGLGAVLMQGGRPVAYMSKTLSERAQAKSVYERELMA 961
T PVLA+PDF KTF LE DASG G+ AVL+QGG P+AY S+ L Y++EL A
Sbjct: 277 TKAPVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYA 456
Query: 962 VVLAVQKWRHYLLGCKFIVHTDQKSLRFLAEQRLMGEEQQKWV 1004
++ A Q W H+L+ +F++H+D +SL+++ + + + KWV
Sbjct: 457 LIRAPQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWV 585
Score = 32.3 bits (72), Expect(2) = 1e-40
Identities = 13/34 (38%), Positives = 18/34 (52%)
Frame = +2
Query: 812 FGQSELIYLGHVISKEGVAADPSKIKDMLNWPLP 845
F + + G V+ + GV DP KIK + WP P
Sbjct: 2 FCVDNIFFSGFVVGRNGVQMDPEKIKAIQEWPPP 103
>BI425021
Length = 426
Score = 163 bits (412), Expect = 6e-40
Identities = 77/142 (54%), Positives = 99/142 (69%)
Frame = -1
Query: 1159 LQPLPIPSQVWSDISMDFIGGLPKTMGKDTILVVVDRFTKYAHFLALSHPYNAKEVAELF 1218
L PLP+P + W D+SMDFI GLP G TI VVV+RF+K H L + A VA LF
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLPPYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASLF 247
Query: 1219 IKEIVKLHGFPTSIVSDRDRVFLSSFWSELFKLAGTKLKFSSAYHPQTDGQTEVVNRCVE 1278
+ ++KLHGFP SIVSDRD +F+S FW +LF+L+GT L+ SSAYHPQTDGQTEV+NR +E
Sbjct: 246 LNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVIE 67
Query: 1279 TYLRCVTGAKPKQWPKWLSWAE 1300
YLR +P+ +++ W E
Sbjct: 66 QYLRAFVHGRPRNLGRFIPWVE 1
>TC213114 weakly similar to UP|Q8W150 (Q8W150) Polyprotein, partial (7%)
Length = 810
Score = 120 bits (302), Expect(2) = 8e-38
Identities = 55/98 (56%), Positives = 75/98 (76%)
Frame = +3
Query: 1365 MKQQANKHRRDIQLKVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEKINAVAYK 1424
MK A++ RR + L VGD VYLK+QPY+LKSLA+++N+KLSPRFYGPY + ++I VA++
Sbjct: 3 MKAYADRSRRAVTLSVGDWVYLKLQPYRLKSLAKKRNEKLSPRFYGPYQIKKQIGLVAFE 182
Query: 1425 LQLPEGSQVHPVFHVSLLKKAPNEGVNSQPLPVYMTEE 1462
L LP ++HPVFH SLLKKA N QPLP+ ++E+
Sbjct: 183 LDLPPARKIHPVFHASLLKKAVAATANPQPLPLMLSED 296
Score = 56.6 bits (135), Expect(2) = 8e-38
Identities = 26/61 (42%), Positives = 37/61 (60%)
Frame = +2
Query: 1463 WELKVEPESIIDSRAGDKGTVEVLVKWKNMPEFENSWEDYTELLEQFPEHHLEDKVILQG 1522
+EL+V P + G EVL++ +++P+FE +WE + EQFP HLEDKV L G
Sbjct: 296 FELRVFPAEVKAVHNNSNGIAEVLIQLEDLPDFEATWESVEVIKEQFPSFHLEDKVTLLG 475
Query: 1523 G 1523
G
Sbjct: 476 G 478
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 155 bits (391), Expect = 2e-37
Identities = 89/254 (35%), Positives = 137/254 (53%), Gaps = 19/254 (7%)
Frame = +1
Query: 637 IEKLVKEMLAAGIIRH-STSPFSSPAILVKKKDG------------------GWRFCVDY 677
+ K V ++L G+I S S + SP ++V KK+G W+ C+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 678 RALNKVTIPDKFPIPIIDELLDEIGTAEIFSKLDLKSGYHQIRMREEDIPKTAFRTHEGH 737
R LN+ T D FP+P +D++L+ + + LD GY+QI + +D K AF G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 738 YEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIYSNNVDLHKEHLREVLQV 797
+ Y +PFGL NAP+TFQ M + + K + VF D ++ +++ + L VLQ
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 798 LRDNHLVANQKKCSFGQSELIYLGHVISKEGVAADPSKIKDMLNWPLPKDVKGLRGFLGL 857
+ +LV N +KC F E I LGH IS G+ D +KI + P P +VKG+R FLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 858 TGYYRRFVRNYSKL 871
+YRRF+++++K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>BQ299538
Length = 426
Score = 108 bits (270), Expect(2) = 3e-36
Identities = 49/101 (48%), Positives = 68/101 (66%)
Frame = +3
Query: 1248 LFKLAGTKLKFSSAYHPQTDGQTEVVNRCVETYLRCVTGAKPKQWPKWLSWAEFWYNTNY 1307
+ KL GT LK S++YHP DGQT VN C+ET+LRC +PK +WLSWAE+WYNTN+
Sbjct: 129 IVKLPGTYLKMSTSYHP*IDGQT--VNHCLETFLRCFVADQPKM*VQWLSWAEYWYNTNF 302
Query: 1308 HSAIKTTPFQALYGREPPVIIKGTDSLASVNEVEKMTAERN 1348
H++ TTPF+ +YGR+PPV+ + V V + +R+
Sbjct: 303 HASTGTTPFEVVYGRKPPVLNRFLPGEVRVEAVRRELQDRD 425
Score = 63.9 bits (154), Expect(2) = 3e-36
Identities = 26/44 (59%), Positives = 35/44 (79%)
Frame = +1
Query: 1205 LSHPYNAKEVAELFIKEIVKLHGFPTSIVSDRDRVFLSSFWSEL 1248
L HPY+A+ +AE+F KE+V LHG P S++SD D +F+SSFW EL
Sbjct: 1 LKHPYSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>AW830191
Length = 372
Score = 148 bits (374), Expect = 2e-35
Identities = 70/123 (56%), Positives = 93/123 (74%)
Frame = +3
Query: 1281 LRCVTGAKPKQWPKWLSWAEFWYNTNYHSAIKTTPFQALYGREPPVIIKGTDSLASVNEV 1340
LRC+TG KP+QWPK LSWAEFW+NTNY++++K TPF+ LYG +PP ++KG ++ EV
Sbjct: 6 LRCLTGTKPQQWPKRLSWAEFWFNTNYNNSLKLTPFKVLYGCDPPHLLKGAIISSTAEEV 185
Query: 1341 EKMTAERNLFLDTLKENLEKAQNRMKQQANKHRRDIQLKVGDMVYLKIQPYKLKSLARRK 1400
MT +R+ L LK NL KAQN+MK A++ RR I L VGD VYLK+QPY+ +SLAR+
Sbjct: 186 NVMTNDRDQMLHDLKGNLAKAQNQMK-YADRSRRSIPLNVGDWVYLKLQPYRQRSLARKT 362
Query: 1401 NQK 1403
N+K
Sbjct: 363 NEK 371
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 148 bits (373), Expect = 2e-35
Identities = 89/254 (35%), Positives = 132/254 (51%), Gaps = 19/254 (7%)
Frame = +1
Query: 637 IEKLVKEMLAAGIIRH-STSPFSSPAILVKKKDG------------------GWRFCVDY 677
+ K V ++L AG+I S S + SP +V KK G WR C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 678 RALNKVTIPDKFPIPIIDELLDEIGTAEIFSKLDLKSGYHQIRMREEDIPKTAFRTHEGH 737
R LN+ T D +P+P +D++L + + LD SGY+QI + +D KTAF
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 738 YEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIYSNNVDLHKEHLREVLQV 797
+ Y +PFGL NA +TFQ M + + K + VF D + + +L +VLQ
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 798 LRDNHLVANQKKCSFGQSELIYLGHVISKEGVAADPSKIKDMLNWPLPKDVKGLRGFLGL 857
++LV N +KC F E I LGH ISK G+ K+ + P P +VKG+ FLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 858 TGYYRRFVRNYSKL 871
G+YRRF+++++K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>CA820403 weakly similar to GP|13273463|gb| pol protein integrase region
{Ginkgo biloba}, partial (52%)
Length = 421
Score = 109 bits (272), Expect(2) = 5e-33
Identities = 53/77 (68%), Positives = 62/77 (79%), Gaps = 1/77 (1%)
Frame = -3
Query: 1218 FIKEIVKLHGFPTSIVSDRDRVFLSS-FWSELFKLAGTKLKFSSAYHPQTDGQTEVVNRC 1276
FIKE VKLHG +SIVSD DR+FL S FW+ELFK+ GTKLKFS AYHPQ D T+VVNRC
Sbjct: 335 FIKEAVKLHGCSSSIVSDWDRLFLIS*FWTELFKMEGTKLKFSLAYHPQPDSHTKVVNRC 156
Query: 1277 VETYLRCVTGAKPKQWP 1293
+E L+C+T +K KQWP
Sbjct: 155 IEMNLQCLTTSKRKQWP 105
Score = 52.0 bits (123), Expect(2) = 5e-33
Identities = 22/34 (64%), Positives = 30/34 (87%)
Frame = -1
Query: 1189 ILVVVDRFTKYAHFLALSHPYNAKEVAELFIKEI 1222
I+V+V RFTKYAHF+ LSHPY AKEV+E+ +K++
Sbjct: 421 IMVIVYRFTKYAHFVVLSHPYLAKEVSEVLLKKL 320
>AW570005
Length = 413
Score = 139 bits (350), Expect = 1e-32
Identities = 67/133 (50%), Positives = 87/133 (65%)
Frame = -2
Query: 843 PLPKDVKGLRGFLGLTGYYRRFVRNYSKLAQPLNQLLKKNNFSWSAGATQAFDKLKEIMT 902
P P+ + LRGFL LTG+YRRF++ Y+ +A PL+ LL K++F WS A AF LK ++T
Sbjct: 412 PPPRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDSFVWSPEADVAFQALKNVVT 233
Query: 903 TVPVLAVPDFQKTFVLETDASGKGLGAVLMQGGRPVAYMSKTLSERAQAKSVYERELMAV 962
VLA+PDF K F +ETDASG +GAVL Q G P+A+ SK + S Y EL A+
Sbjct: 232 NTLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAI 53
Query: 963 VLAVQKWRHYLLG 975
V+KWR YLLG
Sbjct: 52 TNVVKKWRQYLLG 14
>CF922488
Length = 741
Score = 133 bits (334), Expect = 7e-31
Identities = 85/245 (34%), Positives = 126/245 (50%), Gaps = 1/245 (0%)
Frame = +3
Query: 664 VKKKDGGWRFCVDYRALNKVTIPDKFPIPIIDELLDEIGTAEIFSKLDLKSGYHQIRMRE 723
V K+DG CVDYR LN + DKFP+P I+ L+D + FS +D SGY+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 724 EDIPKTAFRTHEGHYEYLVLPFGLTNAPSTFQALMNQVLRPYLRKFVLVFFYDILIYSNN 783
ED+ KT F T G + Y + FGL N +T+Q M + + K + V+ D+++ S
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 784 VDLHKEHLREVLQVLRDNHLVANQKKCSFGQSELIYLGHVISKEGVAADPSKIKDMLNWP 843
+ H +LR++ + LR L N KC F L + S G+ D +K+K +L
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 844 LPKDVKGLRGFLGLTGYYRRFVRNYSKLAQPLNQLLKKNNF-SWSAGATQAFDKLKEIMT 902
P K ++GFLG Y RF+ +PL LL KN F W AF+++K+ +
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 903 TVPVL 907
VL
Sbjct: 723 NPHVL 737
>BI317507
Length = 359
Score = 125 bits (315), Expect = 1e-28
Identities = 61/114 (53%), Positives = 81/114 (70%), Gaps = 1/114 (0%)
Frame = -1
Query: 774 FYDILIYSNNVDLHKEHLREVLQVLRDNHLVANQKKCSFGQSELIYLGHVISKEGVAADP 833
FY+ILIYS + H HL VL VL+ LVAN+KKC F Q+ + YLGHVISK+ VA D
Sbjct: 359 FYNILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDS 180
Query: 834 SKIKDMLNWPLPKDVKGLRGFLGLTGYYRRFVRNYSKLA-QPLNQLLKKNNFSW 886
+K+K ++ WP+PK+VK + FL LTGYYR+F+++Y KLA +PL L K + F W
Sbjct: 179 NKVKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKNDGFKW 18
>BQ627806
Length = 435
Score = 73.2 bits (178), Expect(2) = 1e-26
Identities = 34/83 (40%), Positives = 52/83 (61%)
Frame = +1
Query: 937 PVAYMSKTLSERAQAKSVYERELMAVVLAVQKWRHYLLGCKFIVHTDQKSLRFLAEQRLM 996
P+ + + S Y REL A+ +AV+KWR YLLG F++ TD +SL+ L Q +
Sbjct: 181 PLPFSFMPFCSKLLRASTYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQ 360
Query: 997 GEEQQKWVSKLMGFDFEIKYKPG 1019
EQQ ++++LMGFD+ I+Y+ G
Sbjct: 361 TPEQQIYLARLMGFDYTIQYRAG 429
Score = 67.0 bits (162), Expect(2) = 1e-26
Identities = 31/63 (49%), Positives = 43/63 (68%)
Frame = +3
Query: 878 LLKKNNFSWSAGATQAFDKLKEIMTTVPVLAVPDFQKTFVLETDASGKGLGAVLMQGGRP 937
LL K+ F W+ A +AF +LK + PVL +PDF +FV+ETDASG G+GA+L Q P
Sbjct: 3 LLVKDQFHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHP 182
Query: 938 VAY 940
+A+
Sbjct: 183 LAF 191
>CD487724
Length = 676
Score = 112 bits (281), Expect = 1e-24
Identities = 67/216 (31%), Positives = 114/216 (52%), Gaps = 2/216 (0%)
Frame = +3
Query: 432 ILILVDCGATSNFISQELVAELEIPVVATSEYVVEVGNGARERNSGVCKNLKLEVQGIPI 491
++ LVD G+T NF+ Q LV++L +P +T V VGNG + + +C+ + + +Q I
Sbjct: 24 LVYLVDGGSTHNFVQQPLVSQLGLPCRSTPPLRVMVGNGHHLKCTTICEAIPISIQNIEF 203
Query: 492 IQHFFILGLGGTELVLGMDWLASLGNIEANFQDLIIKWELNGQKMCMQGEPSFCKVAATW 551
+ H ++L + G +VLG+ WL +LG I ++ L +++ + + ++GE
Sbjct: 204 LVHLYVLPIVGANIVLGVQWLKTLGPILVDYNSLSMQFFYQHRLVQLKGESEAQLGLLNH 383
Query: 552 KSIKK--TKHDEGEEYFLSYECSEEEPTANVTIPELWIKLLTEFPEVFQEPKELPPKRAT 609
+++ H+ + ++ PT++ +P+ LL +F +FQ P+ LPP R T
Sbjct: 384 HQLRRLHQTHEPVTYFHIAILTENTSPTSSPPLPQPIQHLLDQFSALFQ*PQGLPPARET 563
Query: 610 DHAILLQEGAPIPNIRPYRYPFYQKNEIEKLVKEML 645
DH I L + N+R Y YP Y NEIE V ML
Sbjct: 564 DHHIHLLP*SEPVNMRLY*YPHY*NNEIEHQVNLML 671
>TC233069
Length = 881
Score = 84.0 bits (206), Expect(2) = 6e-23
Identities = 41/75 (54%), Positives = 51/75 (67%)
Frame = +2
Query: 1131 MKLDIQTYVQKCEICQRNKYETLNPAGYLQPLPIPSQVWSDISMDFIGGLPKTMGKDTIL 1190
M D+ ++ C CQ+NKY T G LQPLPIP QVW DISMDF+ LP + GK I
Sbjct: 224 MARDVCDHICACTNCQQNKYSTQK*FGLLQPLPIP*QVWKDISMDFVTHLPPS*GKKMIW 403
Query: 1191 VVVDRFTKYAHFLAL 1205
V+VD +TKY+HFL+L
Sbjct: 404 VIVDCWTKYSHFLSL 448
Score = 43.5 bits (101), Expect(2) = 6e-23
Identities = 25/71 (35%), Positives = 36/71 (50%)
Frame = +1
Query: 1059 QKIIQEITTQGPVPAGYHMRRGRLLYKNRIVLPKTSGKIPIILQEFHDSAVGGHAGIFRT 1118
QK++Q+ P + G LLY + +P S I+L E HDS +GGH+GI T
Sbjct: 13 QKLLQQKLDNATTP-DCTCQDGLLLYCTHLFIPLESELRAILLGECHDSLLGGHSGIKGT 189
Query: 1119 YKRISALFFWE 1129
R+ F W+
Sbjct: 190 LNRLFVSFAWQ 222
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 97.8 bits (242), Expect(2) = 8e-23
Identities = 53/117 (45%), Positives = 65/117 (55%)
Frame = -2
Query: 1133 LDIQTYVQKCEICQRNKYETLNPAGYLQPLPIPSQVWSDISMDFIGGLPKTMGKDTILVV 1192
L+ + C CQ KYET L PL +P + W D+S+DFI GL ILVV
Sbjct: 353 LECAQMLPNCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPYHVHTAILVV 174
Query: 1193 VDRFTKYAHFLALSHPYNAKEVAELFIKEIVKLHGFPTSIVSDRDRVFLSSFWSELF 1249
VD F+K H L + A VA LFI + KLHG P S+VSD D +F+S FW ELF
Sbjct: 173 VDHFSKGIHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQELF 3
Score = 29.3 bits (64), Expect(2) = 8e-23
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = -1
Query: 1107 SAVGGHAGIFRTYKRISALFFWEGMKLDI 1135
+ GGH GI +T +S +W GM+ D+
Sbjct: 420 TTTGGHTGIAKTLA*LSKNIYWFGMRTDV 334
>BM731326 weakly similar to GP|21740635|em OSJNBb0043H09.2 {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 424
Score = 102 bits (255), Expect = 1e-21
Identities = 51/108 (47%), Positives = 73/108 (67%)
Frame = +1
Query: 1358 LEKAQNRMKQQANKHRRDIQLKVGDMVYLKIQPYKLKSLARRKNQKLSPRFYGPYPVIEK 1417
LE+AQ+ M + AN HRR + VGD VYLKI+P++ S+ R + KL+ RFYGPY V+ +
Sbjct: 25 LERAQSLMVKHANNHRRPHDINVGDWVYLKIRPHRQGSMPPRLHPKLTARFYGPYLVMRQ 204
Query: 1418 INAVAYKLQLPEGSQVHPVFHVSLLKKAPNEGVNSQPLPVYMTEEWEL 1465
+ AVA++LQLP +++HPVFHVS LK+A + LP + + EL
Sbjct: 205 VGAVAFQLQLPSEARIHPVFHVSQLKRALGNHQAQEELPPDLEHQAEL 348
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,785,825
Number of Sequences: 63676
Number of extensions: 957498
Number of successful extensions: 6131
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 5686
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5957
length of query: 1555
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1445
effective length of database: 5,635,272
effective search space: 8142968040
effective search space used: 8142968040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0045.8


