

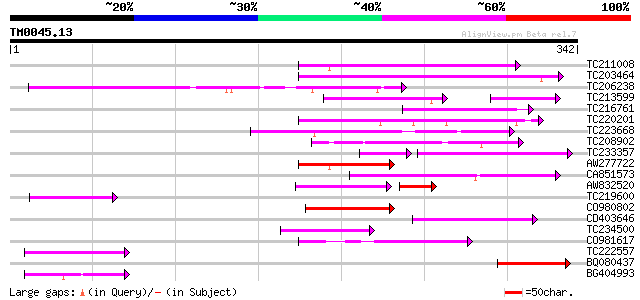
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0045.13
(342 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211008 104 7e-23
TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leish... 97 1e-20
TC206238 weakly similar to UP|Q7PX96 (Q7PX96) AgCP12237 (Fragmen... 88 5e-18
TC213599 homologue to UP|EXBB_PASHA (P72202) Biopolymer transpor... 60 2e-12
TC216761 similar to UP|Q9FQE4 (Q9FQE4) Glutathione S-transferase... 67 9e-12
TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance ... 67 9e-12
TC223668 67 1e-11
TC208902 67 2e-11
TC233357 homologue to UP|Q8VWW2 (Q8VWW2) Two-component response ... 57 3e-10
AW277722 similar to SP|O65719|HS73_ Heat shock cognate 70 kDa pr... 62 3e-10
CA851573 54 1e-07
AW832520 52 1e-07
TC219600 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial ... 53 2e-07
CO980802 52 3e-07
CD403646 52 4e-07
TC234500 52 5e-07
CO981617 50 1e-06
TC222557 50 1e-06
BQ080437 homologue to GP|20197033|gb| expressed protein {Arabido... 50 1e-06
BG404993 49 3e-06
>TC211008
Length = 516
Score = 104 bits (259), Expect = 7e-23
Identities = 55/138 (39%), Positives = 80/138 (57%), Gaps = 4/138 (2%)
Frame = +1
Query: 175 WSPPPPAWVKCNTDGSV----RGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELW 230
W P WV NTDGSV R + +ACGG+ RDS G +L GF NLG ++V AELW
Sbjct: 64 WRCPXNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAELW 243
Query: 231 GIFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCV 290
G+ ++LAWD G ++ V+ DS A+ L+ G P ++S+I + ++W +
Sbjct: 244 GVVHGLKLAWDLGCKKVKVDIDSGNALGLVRHGPVANDPAFALVSEINELVRKEWLVEFS 423
Query: 291 HACREANSVADELANMAH 308
H RE+N AD+LA++ H
Sbjct: 424 HVFRESNRAADKLAHLGH 477
>TC203464 similar to GB|AAF35920.1|7025820|AC005892 L505.8 {Leishmania
major;} , partial (8%)
Length = 1386
Score = 96.7 bits (239), Expect = 1e-20
Identities = 64/165 (38%), Positives = 86/165 (51%), Gaps = 5/165 (3%)
Frame = +2
Query: 175 WSPPPPAWVKCNTDGSVRG-TSNLAACGGVCRDSYGRWLFGFCRNLG-TSNVLWAELWGI 232
W P WVK N DGS S+ A CGGV RD+ +WL GF + L T V EL I
Sbjct: 488 WKKPEIGWVKLNVDGSRDPYKSSSAGCGGVLRDASAKWLRGFAKKLNPTYAVHQTELEAI 667
Query: 233 FSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDR-DWDFQCVH 291
+ +++A + V +L+VESDS V ++ G HP G++ IR + R DW+ + V
Sbjct: 668 LTGLKVASEMNVKKLIVESDSDSVVSMVENGVKPNHPDYGVVELIRTKRRRFDWEVRFVS 847
Query: 292 ACREANSVADELANMAHDLDFGIHILDY--PPPRVSNLLLFDVMG 334
+AN VAD LAN A L +Y PP + LLL D +G
Sbjct: 848 VSNKANRVADRLANDARKLACDDVCEEYLHPPANCNQLLLEDEIG 982
>TC206238 weakly similar to UP|Q7PX96 (Q7PX96) AgCP12237 (Fragment), partial
(9%)
Length = 1259
Score = 88.2 bits (217), Expect = 5e-18
Identities = 71/239 (29%), Positives = 108/239 (44%), Gaps = 11/239 (4%)
Frame = +3
Query: 12 KWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRPL 71
+W GP ++ N L + ++ N R ++DS CP C E ET +H LRDC V
Sbjct: 117 RWVGPERIINLLGNAADGALFTNPLRRYRHTSMDSSCPRCPELEETCLHALRDCPKVAAF 296
Query: 72 WQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRSCFVF-- 129
W+ ++P L F D+ WL L S A FG+ + +W SR+ VF
Sbjct: 297 WRSVLPKKLAPKFFNGDVAVWLETNLSFSEAAFFWPTF---FGIAVELLWESRNDLVFYK 467
Query: 130 QGV--PFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPA----W- 182
G DL + + D + R + +PR + W P P W
Sbjct: 468 DGTWDYLDLSDITDIVFNRYKDCM--RAHASHILMPRNL------LKWRRPLPLSHGHWL 623
Query: 183 VKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLG--TSNVLWAELWGIFSVIQLA 239
++ N G+ +S+ AACGG+ RD+ R++ GF LG SN E+WGI+ +++A
Sbjct: 624 LRLNVSGAYDRSSDTAACGGIFRDNNDRFVLGFSVKLGECLSND-EGEIWGIYHGMKIA 797
>TC213599 homologue to UP|EXBB_PASHA (P72202) Biopolymer transport exbB
protein, partial (8%)
Length = 825
Score = 60.1 bits (144), Expect(2) = 2e-12
Identities = 31/78 (39%), Positives = 46/78 (58%), Gaps = 3/78 (3%)
Frame = +3
Query: 190 SVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVV 249
S+ +++AACGG+ RD +GRW F LG ++ AEL G + + LAWD+G L V
Sbjct: 327 SMHSDNDMAACGGLLRDCHGRWAASFV*KLGNTSAFVAEL*GAYLGLSLAWDKGDKNLEV 506
Query: 250 ESDS---SVAVKLINEGS 264
DS V++K+ +GS
Sbjct: 507 NIDSLVVVVSIKISKQGS 560
Score = 29.6 bits (65), Expect(2) = 2e-12
Identities = 19/42 (45%), Positives = 23/42 (54%)
Frame = +2
Query: 291 HACREANSVADELANMAHDLDFGIHILDYPPPRVSNLLLFDV 332
H REAN AD LAN A ++ IL+ P +S LL DV
Sbjct: 629 HIYREANGCADALANYACAMEEEYVILEQ*PAWLSQLLDSDV 754
>TC216761 similar to UP|Q9FQE4 (Q9FQE4) Glutathione S-transferase GST 14
(Fragment) , complete
Length = 1353
Score = 67.4 bits (163), Expect = 9e-12
Identities = 32/79 (40%), Positives = 47/79 (58%)
Frame = +1
Query: 238 LAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREAN 297
+AW RG+ L+ ESDSS+A++LI+ G + HP +++ IR + D+DW H RE N
Sbjct: 1018 IAWTRGIRNLICESDSSMALQLISTGVYITHPRAALVTTIRSFMDKDWCLSFQHTLREGN 1197
Query: 298 SVADELANMAHDLDFGIHI 316
VAD LA G+H+
Sbjct: 1198 FVADGLAKK------GVHV 1236
>TC220201 similar to UP|Q9ERI5 (Q9ERI5) Apoptotic cell clearance receptor
PtdSerR (Phosphatidylserine receptor), partial (5%)
Length = 1185
Score = 67.4 bits (163), Expect = 9e-12
Identities = 55/163 (33%), Positives = 75/163 (45%), Gaps = 15/163 (9%)
Frame = +1
Query: 175 WSPPPPAWVKCNTDGS-VRGTSNLAACGGVCRDSYGRWLFGFCRNLGTS---NVLWAELW 230
W P WVK N DGS + A CGGV RD +G W GF + L + + EL
Sbjct: 337 WKKPESGWVKLNVDGSRIHEEPASAGCGGVIRDEWGTWCVGFDQKLDPNICRQAHYTELQ 516
Query: 231 GIFSVIQLAWDR--GVPRLVVESDSSVAVKLINE--GSDVLHPYGGMISQI-RQWKDRDW 285
I + +++A + V +LVVESDS AV ++ G P ++ I R KD W
Sbjct: 517 AILTGLKVAREDMINVEKLVVESDSEPAVNMVKSRLGYKYHRPEYKVVQDINRLLKDPKW 696
Query: 286 DFQCVHACREANSVADELA------NMAHDLDFGIHILDYPPP 322
+ + R+AN VA+ LA + HD H YP P
Sbjct: 697 LARLDYVPRKANRVANYLAKRVCKPSSPHDHH---HFSKYPCP 816
>TC223668
Length = 866
Score = 67.0 bits (162), Expect = 1e-11
Identities = 47/161 (29%), Positives = 75/161 (46%), Gaps = 2/161 (1%)
Frame = -3
Query: 146 QQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAW--VKCNTDGSVRGTSNLAACGGV 203
Q + +HC +L + + ++ W+ PPP VK N DGS ++ GG+
Sbjct: 672 QFIKKIHC*GYCLQSSLVSKDLVSSKRLTWTSPPPLLNEVKLNVDGSGHYEPHVMEVGGL 493
Query: 204 CRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINEG 263
RD GRWLF F R+ G N L +EL+ + + ++ +V S + + ++
Sbjct: 492 IRDGVGRWLFRFARHCGLGNPLLSELFALEASLR-------SHIV*TWWSCLLIGQMSR* 334
Query: 264 SDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELA 304
+ +L +I IR + RDW F H + NSV D LA
Sbjct: 333 ACMLMQI--LIHSIRSFMKRDWSFSLCHVNTKVNSVVDFLA 217
>TC208902
Length = 838
Score = 66.6 bits (161), Expect = 2e-11
Identities = 44/130 (33%), Positives = 64/130 (48%), Gaps = 2/130 (1%)
Frame = +1
Query: 183 VKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDR 242
+ N DG+V G +AACG V R+ G + F NLG ++ AE+W I ++LAWD+
Sbjct: 232 INSNCDGAVSG--GVAACGRVLRNQAGAVVV-FAHNLGMCSISQAEIWAITMGVKLAWDK 402
Query: 243 GVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDR--DWDFQCVHACREANSVA 300
G L VESD A+++++ V ++ KD + H REAN A
Sbjct: 403 GFTNLYVESDFKYAIEMMD---GVCEFSNNCFQLVKSAKDSV*GETYSWCHILREANKTA 573
Query: 301 DELANMAHDL 310
D LA D+
Sbjct: 574 DALAKFGVDV 603
>TC233357 homologue to UP|Q8VWW2 (Q8VWW2) Two-component response
regulator-like protein, partial (8%)
Length = 811
Score = 56.6 bits (135), Expect(2) = 3e-10
Identities = 35/93 (37%), Positives = 46/93 (48%)
Frame = +1
Query: 247 LVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANM 306
L VESDS + ++L+ HP G++ I + D A REAN V D AN
Sbjct: 490 LYVESDSLINIQLLKLKYTTTHPAYGIVKMIPDFIDDGVHITWSRALREANQVVDCFANY 669
Query: 307 AHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPR 339
AH L I LD+ P SN+LL D+ + PR
Sbjct: 670 AHSLGVQIRGLDFIPIFASNVLLADLTATAFPR 768
Score = 25.8 bits (55), Expect(2) = 3e-10
Identities = 12/31 (38%), Positives = 16/31 (50%)
Frame = +2
Query: 212 LFGFCRNLGTSNVLWAELWGIFSVIQLAWDR 242
LF F +G + L A+LW I +AW R
Sbjct: 386 LFAFADGIGCFSALNAKLWAXXXGICVAWAR 478
>AW277722 similar to SP|O65719|HS73_ Heat shock cognate 70 kDa protein 3
(Hsc70.3). [Mouse-ear cress] {Arabidopsis thaliana},
partial (8%)
Length = 432
Score = 62.4 bits (150), Expect = 3e-10
Identities = 32/62 (51%), Positives = 39/62 (62%), Gaps = 4/62 (6%)
Frame = +1
Query: 175 WSPPPPAWVKCNTDGSV----RGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELW 230
W PP WV NTDGSV R + +ACGG+ RDS G +L GF NLG ++V AELW
Sbjct: 247 WRCPPNGWVCLNTDGSVFENHRNGCSGSACGGLVRDSSGCYLGGFTVNLGNTSVTLAELW 426
Query: 231 GI 232
G+
Sbjct: 427 GV 432
>CA851573
Length = 614
Score = 53.9 bits (128), Expect = 1e-07
Identities = 41/129 (31%), Positives = 60/129 (45%), Gaps = 2/129 (1%)
Frame = +1
Query: 206 DSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSD 265
DS L F + LG NV+ AELW + I+LA D + E D+ AV +I+
Sbjct: 37 DSIAEXLSCFSKRLGYCNVILAELWSLKYGIKLAIDLNTQFSLFEMDAREAVHMIDXIKS 216
Query: 266 VLHPYGGMISQIRQ--WKDRDWDFQCVHACREANSVADELANMAHDLDFGIHILDYPPPR 323
V + + IR+ W + W VH REANS A+ A H ++ ++D P
Sbjct: 217 VPSSLRPLAADIRRVLWAN-GWQSSIVHINREANSAANWAAKFGHQRNWSSFLVDEIPFV 393
Query: 324 VSNLLLFDV 332
+ L+ DV
Sbjct: 394 LKALVANDV 420
>AW832520
Length = 423
Score = 51.6 bits (122), Expect(2) = 1e-07
Identities = 24/58 (41%), Positives = 32/58 (54%)
Frame = -3
Query: 173 VGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELW 230
+ WSPPP K N DG+V S + GGV RDS+G ++ GF + + AELW
Sbjct: 256 IRWSPPPEGLFKLNYDGAVNCNSLKVSAGGVVRDSHGNFVIGFTTTPDSCTINIAELW 83
Score = 21.6 bits (44), Expect(2) = 1e-07
Identities = 10/22 (45%), Positives = 15/22 (67%)
Frame = -2
Query: 236 IQLAWDRGVPRLVVESDSSVAV 257
+++A G + VESDS+VAV
Sbjct: 71 LRVAHQHGFRSIEVESDSNVAV 6
>TC219600 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial (21%)
Length = 785
Score = 52.8 bits (125), Expect = 2e-07
Identities = 22/53 (41%), Positives = 31/53 (57%)
Frame = +3
Query: 13 WDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDC 65
W+GP K+ F W V G+ N RR +A D +C +C+ +ET +HLLR C
Sbjct: 12 WEGPEKIRLFYWTVAKGGLKTNNRRHHYSLA*DDMCIMCSGAAETYLHLLRAC 170
>CO980802
Length = 803
Score = 52.4 bits (124), Expect = 3e-07
Identities = 23/54 (42%), Positives = 33/54 (60%)
Frame = -3
Query: 179 PPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGI 232
P + K N DGS + ++ AC G+ DS+GR L F N+G N +WA+LWG+
Sbjct: 414 PLLFFKVNVDGSYKASTAKMACRGIILDSFGRVLSCFHSNIGVCNSIWAKLWGL 253
Score = 37.4 bits (85), Expect = 0.010
Identities = 19/67 (28%), Positives = 33/67 (48%), Gaps = 1/67 (1%)
Frame = -2
Query: 64 DCACVRPLWQQLVPGHLFQAFIASDLCPWLLGQLRSS-LLAPIGGGVVPDFGLLLWNIWT 122
DC + W ++P ++ F + L WL L + + AP+G FGL ++++W
Sbjct: 688 DCEDIMKFWSDMIPSNIRAKFFSIGLQDWLAWNLNTKDIKAPLGTWA-SFFGLTVYHLWL 512
Query: 123 SRSCFVF 129
R+ VF
Sbjct: 511 DRTKLVF 491
>CD403646
Length = 580
Score = 52.0 bits (123), Expect = 4e-07
Identities = 26/75 (34%), Positives = 35/75 (46%)
Frame = -1
Query: 244 VPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADEL 303
VP +V + LI + H + MI I+ W +DW H RE N VAD L
Sbjct: 334 VPAFIVSFTGNQWF*LIKGPENSNHQHAAMIEPIKSWLAKDWXVNLSHVWRECNHVADRL 155
Query: 304 ANMAHDLDFGIHILD 318
A H+L G+HI +
Sbjct: 154 AAKGHELPLGLHIFN 110
>TC234500
Length = 866
Score = 51.6 bits (122), Expect = 5e-07
Identities = 23/57 (40%), Positives = 30/57 (52%)
Frame = +1
Query: 164 RRASYHTVQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLG 220
+R Y + V + P W+K N DGS + +L + V RDS G W FGF NLG
Sbjct: 85 KRGHYQMISVE*TFPEVGWIKANVDGSYKALQHLTSSSAVFRDSTGSWCFGFALNLG 255
Score = 37.4 bits (85), Expect = 0.010
Identities = 16/34 (47%), Positives = 24/34 (70%)
Frame = +1
Query: 287 FQCVHACREANSVADELANMAHDLDFGIHILDYP 320
++C H EA +VAD L+ AH++ FG+H+LD P
Sbjct: 637 YRCYH---EATTVADWLSKYAHNIAFGLHVLDVP 729
>CO981617
Length = 837
Score = 50.4 bits (119), Expect = 1e-06
Identities = 33/105 (31%), Positives = 51/105 (48%)
Frame = -1
Query: 175 WSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFS 234
WSPP P K N DG V ++ W+ + ++L A+LW IF
Sbjct: 351 WSPPCPGVSKVNCDGVV-----------FIHEAIATWM-------ASCSILEAKLWSIFY 226
Query: 235 VIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQ 279
++LA D GV +++ESDS AV + E S HP ++ +I++
Sbjct: 225 GMRLAEDCGVQHIIMESDSVEAVNHLMEHSYDNHPLFHLVQEIKR 91
>TC222557
Length = 1002
Score = 50.4 bits (119), Expect = 1e-06
Identities = 22/64 (34%), Positives = 32/64 (49%), Gaps = 1/64 (1%)
Frame = +1
Query: 10 IWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSL-CPLCNEESETIIHLLRDCACV 68
+W+ P K F WR++ I WR + L++L CP CN + E HL +C +
Sbjct: 439 LWQLKIPLKATTFAWRLIKERIPTKGNLWRRRVQLNNLMCPFCNRQEEEASHLFFNCPRI 618
Query: 69 RPLW 72
PLW
Sbjct: 619 LPLW 630
>BQ080437 homologue to GP|20197033|gb| expressed protein {Arabidopsis
thaliana}, partial (13%)
Length = 422
Score = 50.1 bits (118), Expect = 1e-06
Identities = 26/44 (59%), Positives = 30/44 (68%)
Frame = -2
Query: 295 EANSVADELANMAHDLDFGIHILDYPPPRVSNLLLFDVMGGSLP 338
E NSVAD LAN AH+ + GIHI+D+PP LL DV G SLP
Sbjct: 277 EMNSVADYLANEAHNYEVGIHIVDHPPAECRLLLAEDVGGISLP 146
>BG404993
Length = 412
Score = 49.3 bits (116), Expect = 3e-06
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 2/65 (3%)
Frame = -3
Query: 10 IWKWDGPFKVANFLWRVLNNGI--WVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCAC 67
+WK P K F WR++N+ + VN RR + ++ DSLCPLCN E HL +C
Sbjct: 221 LWKLKIPPKAPVFTWRLINDRLPTKVNLRRRQVEIS-DSLCPLCNNSEEDAAHLFFNCNK 45
Query: 68 VRPLW 72
P W
Sbjct: 44 TLPFW 30
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.141 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,838,191
Number of Sequences: 63676
Number of extensions: 454067
Number of successful extensions: 3261
Number of sequences better than 10.0: 133
Number of HSP's better than 10.0 without gapping: 3180
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3226
length of query: 342
length of database: 12,639,632
effective HSP length: 98
effective length of query: 244
effective length of database: 6,399,384
effective search space: 1561449696
effective search space used: 1561449696
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0045.13


