

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
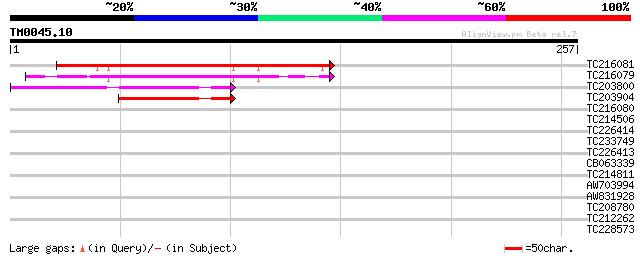
Query= TM0045.10
(257 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216081 similar to UP|Q9ZVA8 (Q9ZVA8) F9K20.6 protein, partial ... 124 6e-29
TC216079 weakly similar to UP|Q9ZVA8 (Q9ZVA8) F9K20.6 protein, p... 111 4e-25
TC203800 weakly similar to UP|Q8WXA1 (Q8WXA1) Inhibitory PAS dom... 93 1e-19
TC203904 67 1e-11
TC216080 weakly similar to GB|CAE76612.1|38567324|BX842641 relat... 40 0.001
TC214506 similar to PIR|T01932|T01932 RNA binding protein homolo... 31 0.49
TC226414 homologue to UP|Q84V95 (Q84V95) Pyruvate decarboxylase ... 30 1.1
TC233749 similar to UP|Q7PTT8 (Q7PTT8) ENSANGP00000015447 (Fragm... 30 1.4
TC226413 homologue to UP|DCP2_PEA (P51851) Pyruvate decarboxylas... 30 1.4
CB063339 30 1.4
TC214811 homologue to UP|VAB2_HORVU (Q40079) Vacuolar ATP syntha... 29 1.8
AW703994 29 2.4
AW831928 28 4.1
TC208780 similar to GB|AAP68270.1|31711828|BT008831 At1g53290 {A... 28 4.1
TC212262 similar to UP|PODK_FLAPR (Q42736) Pyruvate, phosphate d... 28 5.4
TC228573 27 9.2
>TC216081 similar to UP|Q9ZVA8 (Q9ZVA8) F9K20.6 protein, partial (22%)
Length = 1046
Score = 124 bits (310), Expect = 6e-29
Identities = 70/134 (52%), Positives = 88/134 (65%), Gaps = 8/134 (5%)
Frame = +2
Query: 22 SMAVCHRHRSSRATKAT--LTEID-AEHEVTLKMFDDLIQRILVKKATPDWLPFLPGYSF 78
S++V RHRSS+A A + E+D AE++V LK FDDLIQRILVKK TPDWLPF+PG SF
Sbjct: 128 SLSVPLRHRSSKAAAAGGHVVEVDMAEYDVALKHFDDLIQRILVKKNTPDWLPFVPGSSF 307
Query: 79 WVPPRPSPSSVVHLAHRFNSSD--QPQDALNLESH-HGWPDPNYFLQGNAPAHSGESGVE 135
WVPPR SPS+V L H+ + D P D L S GWP N+F+ N ++ +E
Sbjct: 308 WVPPRLSPSNVADLVHKLSYEDHRHPHDCSPLLSTLRGWPSDNFFIDENESTDGEDTDIE 487
Query: 136 LNLPE--EGTVKVK 147
+N P+ GTVKVK
Sbjct: 488 INGPDGTNGTVKVK 529
>TC216079 weakly similar to UP|Q9ZVA8 (Q9ZVA8) F9K20.6 protein, partial (19%)
Length = 538
Score = 111 bits (277), Expect = 4e-25
Identities = 73/145 (50%), Positives = 85/145 (58%), Gaps = 5/145 (3%)
Frame = +3
Query: 8 KVKPLYRNPFPSSNSMAVCHRHRSSRATKATLTEID-AEHEVTLKMFDDLIQRILVKKAT 66
K PL R P S++V R RSS+A A + E+D AE++V LK FDDLIQRILVKK T
Sbjct: 138 KAGPLRRLP-----SLSVPLRRRSSKAA-AAMGEVDMAEYDVALKHFDDLIQRILVKKNT 299
Query: 67 PDWLPFLPGYSFWVPPRPSPSSVVHLAHRFNSSD---QPQDALNLESH-HGWPDPNYFLQ 122
PDWLPF+PG SFWVPPRPS S+V L H+ D P D L S GWP NYF+
Sbjct: 300 PDWLPFVPGSSFWVPPRPSHSNVADLVHKLTYEDHRRHPHDCSPLLSTLRGWPSVNYFID 479
Query: 123 GNAPAHSGESGVELNLPEEGTVKVK 147
+G G GTVKVK
Sbjct: 480 ----EINGLDGT------NGTVKVK 524
>TC203800 weakly similar to UP|Q8WXA1 (Q8WXA1) Inhibitory PAS domain protein,
partial (3%)
Length = 860
Score = 93.2 bits (230), Expect = 1e-19
Identities = 48/102 (47%), Positives = 61/102 (59%)
Frame = +3
Query: 1 MASAFFSKVKPLYRNPFPSSNSMAVCHRHRSSRATKATLTEIDAEHEVTLKMFDDLIQRI 60
MA +K K S+ + VCHRH+SS ATKA EID +K+ ++ +QR+
Sbjct: 63 MARFISNKTKTHLLRHSESAKGVIVCHRHQSSEATKAKWVEIDG-----VKLLEEHVQRV 227
Query: 61 LVKKATPDWLPFLPGYSFWVPPRPSPSSVVHLAHRFNSSDQP 102
+VKKATPDWLPFLPG SFWVPP PSP H+F+ P
Sbjct: 228 MVKKATPDWLPFLPGSSFWVPPPPSP-----FLHKFSIPSHP 338
>TC203904
Length = 911
Score = 66.6 bits (161), Expect = 1e-11
Identities = 29/53 (54%), Positives = 37/53 (69%)
Frame = +2
Query: 50 LKMFDDLIQRILVKKATPDWLPFLPGYSFWVPPRPSPSSVVHLAHRFNSSDQP 102
+++ +D + R++VKKATPDWLPFLPG SFWVPP PSP L H F+ P
Sbjct: 209 VRLLEDHVHRLMVKKATPDWLPFLPGSSFWVPPPPSP-----LLHTFSIPSHP 352
>TC216080 weakly similar to GB|CAE76612.1|38567324|BX842641 related to
deoxyribodipyrimidine photo-lyase {Neurospora crassa;} ,
partial (3%)
Length = 704
Score = 39.7 bits (91), Expect = 0.001
Identities = 44/162 (27%), Positives = 64/162 (39%), Gaps = 20/162 (12%)
Frame = +1
Query: 92 LAHRFNSSDQ--PQDALNLESH-HGWPDPNYFLQGNAPAHSGESGVELNLPE--EGTVKV 146
L H+ + D P D L S GWP N+F+ N ++ +E+N P+ GTVKV
Sbjct: 1 LVHKLSYEDHRHPHDCSPLLSTLRGWPSDNFFIDENESTDGEDTDIEINGPDGTNGTVKV 180
Query: 147 KRVLLWLSDILSVGKRIVNYQLCRSMNYEEHKTPKGIFMVQPKLM--------------E 192
K L S N + +G ++ P + +
Sbjct: 181 K-------------------VLTFSENVAHSEDEEG*SII*PSQLLRFFYDAM*GCGEVQ 303
Query: 193 KSKNTKGKNRGGDREGQFVQMEIPADTALVVVVEL-TLLTLH 233
K K+ K G +GQ VQM+IP+ V + T LTLH
Sbjct: 304 KKKHGKKTKLSGCWKGQCVQMKIPSGYRTVGSFGI*THLTLH 429
>TC214506 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment) {Nicotiana tabacum;} , partial (20%)
Length = 662
Score = 31.2 bits (69), Expect = 0.49
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Frame = +2
Query: 81 PPRPSPSSVVHLAHR--FNSSDQPQDALNLESHHGWPDPNYFLQGNAPAHSGE 131
PP P PSS H H+ SSD+ + + HH W D NY N AH+GE
Sbjct: 395 PPPPPPSSHHHHHHKQAAASSDEIRTVWLGDLHH-WMDENYL--HNCFAHTGE 544
>TC226414 homologue to UP|Q84V95 (Q84V95) Pyruvate decarboxylase 1 , partial
(48%)
Length = 1180
Score = 30.0 bits (66), Expect = 1.1
Identities = 13/40 (32%), Positives = 24/40 (59%)
Frame = -3
Query: 8 KVKPLYRNPFPSSNSMAVCHRHRSSRATKATLTEIDAEHE 47
++ PL+R P+ SS S+ C H+S++ ++ + A HE
Sbjct: 797 QLAPLHRTPWQSSISLHHCGLHQSTQCNSSS*SHCTAHHE 678
>TC233749 similar to UP|Q7PTT8 (Q7PTT8) ENSANGP00000015447 (Fragment),
partial (11%)
Length = 495
Score = 29.6 bits (65), Expect = 1.4
Identities = 13/29 (44%), Positives = 15/29 (50%)
Frame = +2
Query: 91 HLAHRFNSSDQPQDALNLESHHGWPDPNY 119
H AHRFNS D+ L S H D N+
Sbjct: 269 HTAHRFNSLTVRADSFRLRSEHAAADSNF 355
>TC226413 homologue to UP|DCP2_PEA (P51851) Pyruvate decarboxylase isozyme 2
(PDC) (Fragment) , partial (43%)
Length = 735
Score = 29.6 bits (65), Expect = 1.4
Identities = 13/40 (32%), Positives = 25/40 (62%)
Frame = -1
Query: 8 KVKPLYRNPFPSSNSMAVCHRHRSSRATKATLTEIDAEHE 47
++ PL+R P+ SS S+ C H+S++ + ++ + A HE
Sbjct: 387 QLAPLHRTPWQSSISLHHCGLHQSTQCSSSS*SHCMAHHE 268
>CB063339
Length = 380
Score = 29.6 bits (65), Expect = 1.4
Identities = 26/75 (34%), Positives = 31/75 (40%)
Frame = +2
Query: 14 RNPFPSSNSMAVCHRHRSSRATKATLTEIDAEHEVTLKMFDDLIQRILVKKATPDWLPFL 73
+NPFP + S AT A LTE + TLK +L P WL
Sbjct: 20 QNPFPLTTSHV---------ATVAQLTERVPQLFTTLKTLF-----LLPLLMWPPWLNSP 157
Query: 74 PGYSFWVPPRPSPSS 88
G+ PP PSPSS
Sbjct: 158SGFRNTPPPSPSPSS 202
>TC214811 homologue to UP|VAB2_HORVU (Q40079) Vacuolar ATP synthase subunit B
isoform 2 (V-ATPase B subunit 2) (Vacuolar proton pump
B subunit 2) , partial (31%)
Length = 789
Score = 29.3 bits (64), Expect = 1.8
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = +3
Query: 232 LHGLNFGFSFKPVYVEYLQRCYVES 256
LHGL FGFS ++ +LQR + S
Sbjct: 363 LHGLCFGFSLASFFIVFLQRLLISS 437
>AW703994
Length = 411
Score = 28.9 bits (63), Expect = 2.4
Identities = 14/33 (42%), Positives = 20/33 (60%)
Frame = +3
Query: 17 FPSSNSMAVCHRHRSSRATKATLTEIDAEHEVT 49
FPSSN +C R S T+ T +E ++H+VT
Sbjct: 105 FPSSNPACLCVPLRVSACTRHTHSERTSQHKVT 203
>AW831928
Length = 351
Score = 28.1 bits (61), Expect = 4.1
Identities = 15/48 (31%), Positives = 25/48 (51%), Gaps = 1/48 (2%)
Frame = +1
Query: 80 VPPRPSPSSVVHLAHRFNSSDQPQDALN-LESHHGWPDPNYFLQGNAP 126
+ P PS S H +R S+ + ++ ++ HH P P +FLQ + P
Sbjct: 103 IQPPPSSPSPPHFRNRSKPSNPFHNPIH*VQVHHFLPAPLHFLQQHXP 246
>TC208780 similar to GB|AAP68270.1|31711828|BT008831 At1g53290 {Arabidopsis
thaliana;} , partial (51%)
Length = 969
Score = 28.1 bits (61), Expect = 4.1
Identities = 18/79 (22%), Positives = 34/79 (42%)
Frame = -1
Query: 90 VHLAHRFNSSDQPQDALNLESHHGWPDPNYFLQGNAPAHSGESGVELNLPEEGTVKVKRV 149
++ H+F SS + L ++ W +F + P H G S +++ + + +
Sbjct: 564 LYYTHKFYSSSDSRVGLLEQASF*WSSNIFFSGLHRPEHFGISHTAIDVAVQSVEQSS*L 385
Query: 150 LLWLSDILSVGKRIVNYQL 168
WL+ I S+ IV L
Sbjct: 384 FSWLTFIASIQAPIVTSSL 328
>TC212262 similar to UP|PODK_FLAPR (Q42736) Pyruvate, phosphate dikinase,
chloroplast precursor (Pyruvate, orthophosphate
dikinase) , partial (17%)
Length = 983
Score = 27.7 bits (60), Expect = 5.4
Identities = 11/23 (47%), Positives = 14/23 (60%)
Frame = -3
Query: 69 WLPFLPGYSFWVPPRPSPSSVVH 91
WLPFLPG ++P R S +H
Sbjct: 426 WLPFLPGLLLFLPTRTSWPCFLH 358
>TC228573
Length = 1628
Score = 26.9 bits (58), Expect = 9.2
Identities = 13/30 (43%), Positives = 18/30 (59%)
Frame = -1
Query: 186 VQPKLMEKSKNTKGKNRGGDREGQFVQMEI 215
V PKL + +G+NR GD G+FV +I
Sbjct: 815 VVPKLQVRQFRRQGRNRVGDIPGKFVVRKI 726
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,236,893
Number of Sequences: 63676
Number of extensions: 181924
Number of successful extensions: 1237
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 1226
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1234
length of query: 257
length of database: 12,639,632
effective HSP length: 95
effective length of query: 162
effective length of database: 6,590,412
effective search space: 1067646744
effective search space used: 1067646744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0045.10


