

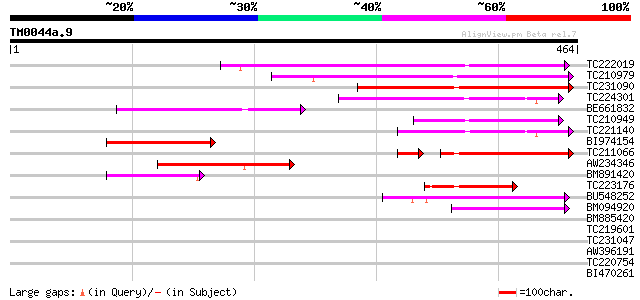
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0044a.9
(464 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC222019 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-li... 185 4e-47
TC210979 142 4e-34
TC231090 125 4e-29
TC224301 weakly similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, par... 106 2e-23
BE661832 91 1e-18
TC210949 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-li... 86 4e-17
TC221140 similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (20%) 78 7e-15
BI974154 77 1e-14
TC211066 69 1e-14
AW234346 75 6e-14
BM891420 similar to GP|20197120|gb| expressed protein {Arabidops... 68 8e-12
TC223176 weakly similar to UP|Q8LPF4 (Q8LPF4) AT5g02170/T7H20_22... 50 2e-06
BU548252 similar to GP|15290010|db P0683F02.7 {Oryza sativa (jap... 42 4e-04
BM094920 weakly similar to GP|6579208|gb| T23G18.9 {Arabidopsis ... 42 8e-04
BM885420 38 0.008
TC219601 similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial (23%) 38 0.008
TC231047 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial ... 37 0.019
AW396191 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsi... 37 0.024
TC220754 similar to UP|Q85V22 (Q85V22) Histidine amino acid tran... 36 0.032
BI470261 similar to GP|21554158|gb amino acid permease-like prot... 36 0.042
>TC222019 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-like, partial
(84%)
Length = 979
Score = 185 bits (469), Expect = 4e-47
Identities = 104/288 (36%), Positives = 162/288 (56%), Gaps = 2/288 (0%)
Frame = +1
Query: 173 DNLTTVLHLKQLHLA--KLSAPQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIF 230
DNLT + L L +L + + + A L+ +P++WL+DL IS LS+ GV +++I
Sbjct: 43 DNLTGLFPGTSLDLGSFRLDSVHLFGILAALIIIPTVWLKDLRIISILSAGGVFATLLIV 222
Query: 231 VSVAATAVFGGVRVNHEIPFLRLHNIPSISGIYIFSYGGHIVFPNLYKAMKDPSKFTKVS 290
V V GV +H + IP GI+ F + GH VFPN+Y++M D +FTK
Sbjct: 223 VCVFCVGTINGVGFHHTGQLVNWSGIPLAIGIHGFCFAGHSVFPNIYQSMADKRQFTKAL 402
Query: 291 ILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMTKYALEF 350
I+ F +Y + MG MFG E SQ+TL+MP +K+ALW TV+ P TKYAL
Sbjct: 403 IICFVLSITIYGGVAIMGFLMFGGETLSQITLNMPRDAFASKVALWTTVINPFTKYALLM 582
Query: 351 APVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAI 410
P+A LE LP +S + + ++ ++L++ + A +P+F V++L GSL SV +
Sbjct: 583 NPLARSLEELLPDRISSTYRCFI--LLRTALVVSTVCAAFLIPFFGFVMALIGSLFSVLV 756
Query: 411 SLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLV 458
S+I P ++KI + + L++ I TFG + G++GT SS + +V
Sbjct: 757 SVIMPSLCFMKIVGKKATATQVALSVVITTFGVICGILGTYSSVQNIV 900
>TC210979
Length = 928
Score = 142 bits (357), Expect = 4e-34
Identities = 78/249 (31%), Positives = 142/249 (56%), Gaps = 2/249 (0%)
Frame = +2
Query: 215 ISFLSSVGVLMSVVIFVSVAATAVFGGVRVNHE--IPFLRLHNIPSISGIYIFSYGGHIV 272
+S++S+ GV+ ++++ + + + ++ + L P G+Y + Y GH V
Sbjct: 2 LSYISACGVIATILVVLCLFWVGLLDNADIHTQGTTKTFNLATFPVAIGLYGYCYAGHAV 181
Query: 273 FPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTK 332
F N+Y AM + ++F V ++ F T++Y + MG FG SQ TL+MP + TK
Sbjct: 182 FSNIYTAMANRNQFPGVLLVCFAICTSMYCAVAIMGYTAFGKATLSQYTLNMPQHLVATK 361
Query: 333 IALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTV 392
IA+W TV+ P TKYAL +PVA+ LE +P+ + I +I ++L++ L + L+V
Sbjct: 362 IAVWTTVVNPFTKYALSLSPVAMCLEELIPA--NSPNFFIYSKLIRTALVVSTLLVGLSV 535
Query: 393 PYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTIS 452
P V+SLTGSL+++ +SLI P A ++ I G+I++ + ++II G + +G+ S
Sbjct: 536 PXXGLVMSLTGSLLTMFVSLILPAACFLSIRGGRITRFQVSICVTIIVVGIVSSCLGSYS 715
Query: 453 SSKLLVEKI 461
+ +++++
Sbjct: 716 ALSEIIQEL 742
>TC231090
Length = 559
Score = 125 bits (314), Expect = 4e-29
Identities = 70/178 (39%), Positives = 108/178 (60%), Gaps = 1/178 (0%)
Frame = +2
Query: 285 KFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQIVTKIALWATVLTPMT 344
+F V + F T LY +G MFG + SQ TL+MP + + TKIA+W TV+ P T
Sbjct: 2 QFPGVLLACFGICTLLYAGAAVLGYTMFGEAILSQFTLNMPKELVATKIAVWTTVVNPFT 181
Query: 345 KYALEFAPVAIQLEHKLPSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHVLSLTG 403
KYAL +PVA+ LE +PS+ K L I I + L+L L + L+VP+F V+SL G
Sbjct: 182 KYALTISPVAMSLEELIPSN---HAKSYLYSIFIRTGLVLSTLVIGLSVPFFGLVMSLIG 352
Query: 404 SLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
SL+++ ++LI PCA +++I RG++++ L I+IIT G + G+ S+ +V+ +
Sbjct: 353 SLLTMLVTLILPCACFLRILRGKVTRTQAALCITIITVGVVCSAFGSYSALAEIVKSL 526
>TC224301 weakly similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (29%)
Length = 667
Score = 106 bits (265), Expect = 2e-23
Identities = 60/186 (32%), Positives = 108/186 (57%), Gaps = 2/186 (1%)
Frame = +2
Query: 270 HIVFPNLYKAMKDPSKFTKVSILSFTTVTALYTIMGYMGAKMFGPEVNSQVTLSMPSKQI 329
H + Y +M+D S+F++ + F+ Y G + MFG EV SQVTL++P+ +
Sbjct: 2 HPIXXTXYNSMRDKSQFSRXXSICFSVCXLGYAAAGVLXYLMFGQEVESQVTLNLPTGKF 181
Query: 330 VTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALA 389
+ +A++ T++ P+TKYAL PV +++K+ + R+ + + +S+L+ L +A
Sbjct: 182 SSHVAIFTTLVNPITKYALMLTPVIYAVKNKVSWHYNKRSTHM---FVITSMLISTLIVA 352
Query: 390 LTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISK--PLFVLNISIITFGFLLGV 447
+ +P F +++SL G+L+SV+ S++ P Y+KI G + ++N SII G + V
Sbjct: 353 VAIPLFGYLMSLIGALLSVSASILVPSVCYLKI-SGAYKRFGSEMIINYSIIIMGVTIAV 529
Query: 448 VGTISS 453
VGT +S
Sbjct: 530 VGTYTS 547
>BE661832
Length = 635
Score = 90.5 bits (223), Expect = 1e-18
Identities = 52/155 (33%), Positives = 86/155 (54%)
Frame = +1
Query: 88 GLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKLRSYVDIGNHAFGAK 147
G+ +S PY + GGW S LL A+ YT L+ + + K+ +R+Y D+G AFG
Sbjct: 145 GINAVSVPYTLASGGWLSLALLFAIAAATFYTGVLIKR*MDKDSNIRTYPDMGELAFGKT 324
Query: 148 GRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHLKQLHLAKLSAPQILTVGAVLVALPSL 207
GR + + LIY ++F+ V + I DNL+ + ++H A L+ + + V L +L
Sbjct: 325 GRLIISGLIYTELFLVSVGFLILEGDNLSNLFPTVEIHTADLA----IGGKKLFVILVAL 492
Query: 208 WLRDLSSISFLSSVGVLMSVVIFVSVAATAVFGGV 242
L +L +S++S+ V S +I +S++ TA F GV
Sbjct: 493 VLDNLRILSYVSASRVFASAIIILSISWTATFDGV 597
>TC210949 similar to UP|Q6K4N7 (Q6K4N7) Amino acid transporter-like, partial
(35%)
Length = 569
Score = 85.9 bits (211), Expect = 4e-17
Identities = 45/123 (36%), Positives = 74/123 (59%)
Frame = +2
Query: 331 TKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALAL 390
+K+ALW TV+ P+TKYAL P+A LE LP +S + ++ ++L+ + +A
Sbjct: 14 SKVALWTTVINPLTKYALLMNPLARSLEELLPDRISSSYWCFM--LLRTTLVASTVCVAF 187
Query: 391 TVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGT 450
VP+F V++L GSL S+ +S I P ++KI + +K L+++I FG + G++GT
Sbjct: 188 LVPFFGLVMALIGSLFSILVSAIMPSLCFLKIIGKKATKTQVALSVAIAAFGVICGILGT 367
Query: 451 ISS 453
SS
Sbjct: 368 YSS 376
>TC221140 similar to UP|Q9MBG9 (Q9MBG9) Gb|AAC79623.2, partial (20%)
Length = 644
Score = 78.2 bits (191), Expect = 7e-15
Identities = 45/146 (30%), Positives = 85/146 (57%), Gaps = 2/146 (1%)
Frame = +1
Query: 318 SQVTLSMPSKQIVTKIALWATVLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGII 377
SQVTL++P ++ +K+A++ T++ P++K+AL P+ L+ LP + R IL +
Sbjct: 10 SQVTLNLPLNKVSSKLAIYTTLVNPISKFALMATPITNALKDLLPRAYKNRATNIL---V 180
Query: 378 GSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISK--PLFVLN 435
+ L++ +AL+VP+F ++SL G+ +SV S++ PC Y+KI G ++ +
Sbjct: 181 STVLVISATIVALSVPFFGDLMSLVGAFLSVTASILLPCLCYLKI-SGTYNEFGCETIAI 357
Query: 436 ISIITFGFLLGVVGTISSSKLLVEKI 461
++II +G+ GT +S +V +
Sbjct: 358 VTIIVAAIAMGISGTYTSVMEIVHHL 435
>BI974154
Length = 420
Score = 77.4 bits (189), Expect = 1e-14
Identities = 34/89 (38%), Positives = 56/89 (62%)
Frame = +2
Query: 80 MNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKLRSYVDI 139
+N + +L G+G LSTPYA + GGW +L+ ++ YT LL CL P+L +Y DI
Sbjct: 2 LNGINVLCGVGILSTPYAAKVGGWLGLSILVIFAIISFYTGLLLRSCLDSEPELETYPDI 181
Query: 140 GNHAFGAKGRFLAAILIYMDIFMSLVSYT 168
G AFG GR +I++Y+++++S +++
Sbjct: 182 GQAAFGTTGRIAISIVLYVELYVSTDAFS 268
>TC211066
Length = 805
Score = 68.9 bits (167), Expect(2) = 1e-14
Identities = 41/110 (37%), Positives = 68/110 (61%), Gaps = 1/110 (0%)
Frame = +2
Query: 353 VAIQLEHKLPSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHVLSLTGSLVSVAIS 411
VA+ LE +PS+ K L I I + L+L L + L+VP+F V+SL GSL+++ ++
Sbjct: 116 VAMSLEELIPSN---HAKSYLYSIFIRTGLVLSTLFIGLSVPFFGLVMSLIGSLLTMLVT 286
Query: 412 LIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEKI 461
LI PCA +++I RG++++ L I+IIT G + GT S+ +V+ +
Sbjct: 287 LILPCACFLRILRGKVTRIQAALCITIITVGVVCSAFGTYSALSEIVKSL 436
Score = 28.5 bits (62), Expect(2) = 1e-14
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +3
Query: 318 SQVTLSMPSKQIVTKIALWAT 338
SQ TL+MP + + T IA+W T
Sbjct: 54 SQFTLNMPKELVATNIAVWTT 116
>AW234346
Length = 371
Score = 75.1 bits (183), Expect = 6e-14
Identities = 40/114 (35%), Positives = 69/114 (60%), Gaps = 2/114 (1%)
Frame = +2
Query: 122 LLGKCLKKNPKLRSYVDIGNHAFGAKGRFLAAILIYMDIFMSLVSYTISLHDNLTTVLHL 181
L+ +C+ +P ++++ DIG AFG KGR + +I + ++F+ + + I DNL ++
Sbjct: 11 LVKRCMDMDPDIKNFPDIGQRAFGDKGRIIVSIAMNSELFLVVTGFLILEGDNLNKLVPN 190
Query: 182 KQLHLAKLS--APQILTVGAVLVALPSLWLRDLSSISFLSSVGVLMSVVIFVSV 233
QL LA L+ I T+ A LV LP++ L DLS +S++S+ G L S + +S+
Sbjct: 191 MQLELAGLTIGGTTIFTMIAALVILPTVLLEDLSMLSYVSASGALASSIFLLSI 352
>BM891420 similar to GP|20197120|gb| expressed protein {Arabidopsis
thaliana}, partial (12%)
Length = 433
Score = 68.2 bits (165), Expect = 8e-12
Identities = 31/84 (36%), Positives = 49/84 (57%), Gaps = 4/84 (4%)
Frame = +2
Query: 80 MNMVGMLIGLGQLSTPYAVEQGGWASAILLIALGVMCTYTCHLLGKCLKKNPKLRSYVDI 139
++ ++ G+G LSTPY +++ GW S +L++ V+C YT L+ C + + SY DI
Sbjct: 164 LSATNVMAGVGILSTPYTLKEAGWMSMVLMVLFAVICCYTATLMRYCFESREGITSYPDI 343
Query: 140 GNHAFGAKGRFLA----AILIYMD 159
G AFG GR + +I IY+D
Sbjct: 344 GEAAFGKYGRIIVSVSLSIFIYID 415
>TC223176 weakly similar to UP|Q8LPF4 (Q8LPF4) AT5g02170/T7H20_220, partial
(11%)
Length = 437
Score = 50.1 bits (118), Expect = 2e-06
Identities = 33/77 (42%), Positives = 49/77 (62%), Gaps = 1/77 (1%)
Frame = +2
Query: 340 LTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGI-IGSSLLLVILALALTVPYFEHV 398
LT +T YAL +PVA+ LE +PS+ K L I I + L+L L + L+VP+F V
Sbjct: 218 LTSIT-YALTISPVAMSLEELIPSN---HAKSYLYSIFIRTGLVLSTLFIGLSVPFFGLV 385
Query: 399 LSLTGSLVSVAISLIFP 415
+SL GSL+++ ++LI P
Sbjct: 386 MSLIGSLLTMLVTLILP 436
>BU548252 similar to GP|15290010|db P0683F02.7 {Oryza sativa (japonica
cultivar-group)}, partial (45%)
Length = 671
Score = 42.4 bits (98), Expect = 4e-04
Identities = 36/160 (22%), Positives = 69/160 (42%), Gaps = 7/160 (4%)
Frame = -3
Query: 306 YMGAKMFGPEVNSQVTLSMPSKQ--IVTKIALWATV----LTPMTKYALEFAPVAIQLEH 359
Y+G GPE + + ++ I IA+ AT + P + L+ ++
Sbjct: 501 YIGDSSKGPEKDYSLKGDSVNRLFGIFNAIAIIATTYGNGIIPEIQVYLQPTNEVLEQTF 322
Query: 360 KLPSSMSGRTKMILRGIIGSSLLLVI-LALALTVPYFEHVLSLTGSLVSVAISLIFPCAF 418
P S + ++ +I SL + I +A +P+F + SL G+ + + I P F
Sbjct: 321 GDPKSPEFSKRNVIPRVISRSLAIAISTTIAAMLPFFGDINSLIGAFGFIPLDFILPMVF 142
Query: 419 YIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLV 458
Y + P+F LN+ I+ LG + +++ + +V
Sbjct: 141 YNLTFKPSKRSPVFWLNVIIVVAFSALGAIAAVAAVRQIV 22
>BM094920 weakly similar to GP|6579208|gb| T23G18.9 {Arabidopsis thaliana},
partial (31%)
Length = 425
Score = 41.6 bits (96), Expect = 8e-04
Identities = 24/98 (24%), Positives = 46/98 (46%), Gaps = 1/98 (1%)
Frame = +3
Query: 362 PSSMSGRTKMILRGIIGSSLLLVI-LALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYI 420
P S + ++ +I SL + I +A +P+F + SL G+ + + I P FY
Sbjct: 126 PKSPEFSKRNVIPRVISRSLAIAISTTIAAMLPFFGDINSLIGAFGFIPLDFILPMVFYN 305
Query: 421 KICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLV 458
+ P+F LN+ I+ LG + +++ + +V
Sbjct: 306 LTFKPSKRSPVFWLNVIIVVAFSALGAIAAVAAVRQIV 419
>BM885420
Length = 421
Score = 38.1 bits (87), Expect = 0.008
Identities = 14/32 (43%), Positives = 27/32 (83%)
Frame = +1
Query: 398 VLSLTGSLVSVAISLIFPCAFYIKICRGQISK 429
V+SL GSL+++ ++LI PCA +++I RG++++
Sbjct: 232 VMSLIGSLLTMLVTLILPCACFLRILRGKVTR 327
>TC219601 similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial (23%)
Length = 693
Score = 38.1 bits (87), Expect = 0.008
Identities = 22/90 (24%), Positives = 45/90 (49%), Gaps = 1/90 (1%)
Frame = +3
Query: 45 KCDHINNIEESAVARDTNVDAEHDAKANSSLAHSVMNMVGMLIGLGQLSTPYAVEQGGWA 104
K ++ N +E+ +D + A K+ S H ++ ++ LS PYA GW
Sbjct: 255 KSENPNALEQLQHQKDVDAGALFVLKSKGSWMHCGYHLTTSIVAPPLLSLPYAFTFLGWT 434
Query: 105 SAILLIALGVMCT-YTCHLLGKCLKKNPKL 133
+ IL + +G + T Y+ +L+ + L+ + ++
Sbjct: 435 AGILSLVIGALVTFYSYNLISRVLEHHAQM 524
>TC231047 weakly similar to UP|Q9SGD7 (Q9SGD7) T23G18.9, partial (31%)
Length = 522
Score = 37.0 bits (84), Expect = 0.019
Identities = 21/88 (23%), Positives = 43/88 (48%)
Frame = +2
Query: 371 MILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAISLIFPCAFYIKICRGQISKP 430
++ R ++ S + LA +P+F +++L G+ + + I P FY + +
Sbjct: 155 VVPRVVLRSLSVAAFTVLAAMLPFFPDIMALFGAFGCIPLDFILPMVFYNMTFKPSKNTI 334
Query: 431 LFVLNISIITFGFLLGVVGTISSSKLLV 458
+F +N I +L V+G I+S + +V
Sbjct: 335 MFWVNNVIAVASSILVVIGGIASIRQIV 418
>AW396191 similar to GP|14194151|gb AT5g40780/K1B16_3 {Arabidopsis thaliana},
partial (29%)
Length = 444
Score = 36.6 bits (83), Expect = 0.024
Identities = 23/120 (19%), Positives = 50/120 (41%)
Frame = -3
Query: 339 VLTPMTKYALEFAPVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHV 398
V+ + Y L PV +E + + + LR ++ + + + + +T P+F +
Sbjct: 430 VIHVIGSYQLYAMPVFDMIETVMVKQLRFKPTWQLRFVVRNVYVAFTMFVGITFPFFGAL 251
Query: 399 LSLTGSLVSVAISLIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLV 458
L G + PC ++ I + + ++ N I FG LL ++ I + ++
Sbjct: 250 LGFFGGFAFAPTTYFLPCIIWLAIYKPKKFSLSWITNWICIIFGLLLMILSPIGGLRSII 71
>TC220754 similar to UP|Q85V22 (Q85V22) Histidine amino acid transporter,
partial (27%)
Length = 664
Score = 36.2 bits (82), Expect = 0.032
Identities = 20/109 (18%), Positives = 47/109 (42%)
Frame = +1
Query: 352 PVAIQLEHKLPSSMSGRTKMILRGIIGSSLLLVILALALTVPYFEHVLSLTGSLVSVAIS 411
PV +E + ++ +LR ++ + + + +A+T P+F+ +L G +
Sbjct: 28 PVFDMIETVMVKKLNFEPSRMLRFVVRNVYVAFTMFIAITFPFFDGLLGFFGGFAFAPTT 207
Query: 412 LIFPCAFYIKICRGQISKPLFVLNISIITFGFLLGVVGTISSSKLLVEK 460
PC ++ I + + + +N I G L ++ I + ++ K
Sbjct: 208 YFLPCIMWLAIHKPKRYSLSWFINWICIVLGLCLMILSPIGGLRTIIIK 354
>BI470261 similar to GP|21554158|gb amino acid permease-like protein
{Arabidopsis thaliana}, partial (26%)
Length = 421
Score = 35.8 bits (81), Expect = 0.042
Identities = 27/102 (26%), Positives = 47/102 (45%), Gaps = 11/102 (10%)
Frame = +2
Query: 66 EHDAKANSSLA------HSVMNMVGMLIGLGQLSTPYAVEQGGWASAIL-LIALGVMCTY 118
E DA AN L H+ ++ ++G L+ PYA+ GW + L A+G++ Y
Sbjct: 62 EKDAGANFVLQSKGEWWHAGFHLTTAIVGPTILTLPYALRGLGWGLGLFCLTAMGLVTFY 241
Query: 119 TCHLLGK----CLKKNPKLRSYVDIGNHAFGAKGRFLAAILI 156
+ +L+ K C + + ++ H FG+ + ILI
Sbjct: 242 SYYLMSKVLYHCENAGRRHIRFRELAAHVFGSGWMYYFVILI 367
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,720,193
Number of Sequences: 63676
Number of extensions: 300505
Number of successful extensions: 2184
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 2162
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2177
length of query: 464
length of database: 12,639,632
effective HSP length: 101
effective length of query: 363
effective length of database: 6,208,356
effective search space: 2253633228
effective search space used: 2253633228
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0044a.9


