

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
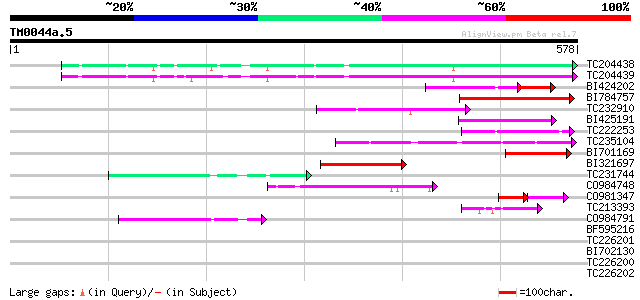
Query= TM0044a.5
(578 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 107 1e-23
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 106 2e-23
BI424202 75 4e-22
BI784757 97 3e-20
TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial ... 87 2e-17
BI425191 weakly similar to GP|14586969|gb| pol polyprotein {Citr... 84 2e-16
TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement p... 82 7e-16
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 77 2e-14
BI701169 72 5e-13
BI321697 68 1e-11
TC231744 56 5e-08
CO984748 53 4e-07
CO981347 39 9e-07
TC213393 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 43 4e-04
CO984791 43 4e-04
BF595216 40 0.002
TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 40 0.004
BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea m... 39 0.006
TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 39 0.008
TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp... 39 0.008
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 107 bits (267), Expect = 1e-23
Identities = 127/556 (22%), Positives = 223/556 (39%), Gaps = 30/556 (5%)
Frame = +1
Query: 53 ELKTERKKRREDELLCRGHIMNTLSDRLYDLYTDTQSAAKIWKTLEFKFKAEEEGIKKFL 112
E T KK+R+ +CR +T S++ D D + +++ E + E L
Sbjct: 940 ECPTHLKKQRKGLSVCRSD--DTESEQESDSDRDVNALTGRFESAEDSSDTDSEITFDEL 1113
Query: 113 ISKYFDFKMLDTKPILQQVHELQVLVNKIKAVK-------IDIRGAFQVGAIIAKLPPSW 165
Y + + ++ ILQQ +L+ ++ ++A K +++G +VG + +KL
Sbjct: 1114 AISYREL-CIKSEKILQQEAQLKKVIANLEAEKEAHEEEISELKG--EVGFLNSKLENMT 1284
Query: 166 NGYRKKLLHNYEDFSLEKIQKHLRIEEESKVRDKAESSG------FSKANTVTTKGKKKY 219
K+L+ D E +Q + + + +S+G F A T G
Sbjct: 1285 KSI--KMLNKGSDMLDEVLQLGKNVGNQRGLGFNHKSAGRTTMTEFVPAKNST--GATMS 1452
Query: 220 DGKQQHLGPKKEHNKFKNNNGTKGPKGGCYVCGKPDHFARDC----------RQNKTKKE 269
+ +H G +++ +K K K C+ CGK H C Q+ +
Sbjct: 1453 QHRSRHHGTQQKKSKRK--------KWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSSSGR 1608
Query: 270 VNAVQVDDEIIATVSEVMAVKGKVPGWWYDTCASVHVSYDKASFKTYSESTDDQEVQMRN 329
+I++ V W+ D+ S H++ K F E V +
Sbjct: 1609 KMMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVK-EFLVNIEPCSTSYVTFGD 1785
Query: 330 EVRSKVVGTGTVELNFTSGKKVTLVNVLHVPEMSRDLVSGDLLGKPGIKSVY-ESEKLIL 388
+ K+ G G + +L VL V ++ +L+S L G + +SE L+
Sbjct: 1786 GSKGKITGMGKL----VHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVT 1953
Query: 389 TRNGVFVGKGYSAEGMIKLCPIDNIINKVSNSAYMIDSVSLWHSRLAHIGISTMNRLIKS 448
+ KG ++ L + D V +WH R H+ + M ++I
Sbjct: 1954 NEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDK 2133
Query: 449 KLI----SCNIHEFEKCEICVKSKMIKKHFKSVERKFN--LLDLVHSDLCEFNGMLTRGG 502
+ + I E C C K +K + ++ + +L+L+H DL + + GG
Sbjct: 2134 GAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGG 2313
Query: 503 NRYFITFIDDCSRYTHVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGEYFSTK 562
RY +DD SR+T V ++ K D F FK ++ + + IK +RSD G E+ ++K
Sbjct: 2314 KRYAYVVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSK 2493
Query: 563 FDSFCEEHDIIHECSA 578
F FC I HE SA
Sbjct: 2494 FTEFCTSEGITHEFSA 2541
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 106 bits (265), Expect = 2e-23
Identities = 127/560 (22%), Positives = 231/560 (40%), Gaps = 34/560 (6%)
Frame = +1
Query: 53 ELKTERKKRREDELLCRGHIMNTLSDRLYDLYTDTQSAAKIWKTLEFKFKAEEEGIKKFL 112
E T KK R+ +C+ +T S++ D D + I++T E + E L
Sbjct: 940 ECPTHLKKHRKGLSVCQS---DTESEQESDSDRDVNALTGIFETAEDSSDTDSEITFDEL 1110
Query: 113 ISKYFDFKMLDTKPILQQVHELQVLVNKIKAVK-------IDIRGAFQVGAIIAKLPPSW 165
+ Y + ++ ILQQ +L+ ++ ++A K +++G +VG + +KL
Sbjct: 1111 AASYRKL-CIKSEKILQQEAQLKKVIADLEAEKEAHKEEISELKG--EVGFLNSKLENMT 1281
Query: 166 NGYRKKLLHNYEDFSLEKI---------QKHLRIEEESKVRDKAESSGFSKANTVTTKGK 216
K+L+ D +L+++ Q+ L +S R +K T T +
Sbjct: 1282 KSI--KMLNKGSD-TLDEVLLLGKNAGNQRGLGFNPKSAGRTTMTEFVPAKNRTGATMSQ 1452
Query: 217 KKYDGKQQHLGPKKEHNKFKNNNGTKGPKGGCYVCGKPDHFARDC-----------RQNK 265
+ +H G +++ +K K K C+ CGK H C + +
Sbjct: 1453 HR----SRHHGMQQKKSKRK--------KWRCHYCGKYGHIKPFCYHLHGHPHHGTQSSN 1596
Query: 266 TKKEVNAVQVDDEIIATVSEVMAVKGKVPGWWYDTCASVHVSYDKASFKTYSESTDDQEV 325
++K++ V + V + K W+ D+ S H++ K F E V
Sbjct: 1597 SRKKMMWVPKHKAVSLVVHTSLRASAK-EDWYLDSGCSRHMTGVK-EFLLNIEPCSTSYV 1770
Query: 326 QMRNEVRSKVVGTGTVELNFTSGKKVTLVNVLHVPEMSRDLVSGDLLGKPGIKSVY-ESE 384
+ + K++G G + +L VL V ++ +L+S L G + +SE
Sbjct: 1771 TFGDGSKGKIIGMGKL----VHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSE 1938
Query: 385 KLILTRNGVFVGKGYSAEGMIKLCPIDNIINKVSNSAYMIDSVSLWHSRLAHIGISTMNR 444
L+ + KG ++ L + + D V +WH R H+ + M +
Sbjct: 1939 CLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKK 2118
Query: 445 LIKSKLI----SCNIHEFEKCEICVKSKMIKKHFKSVERKFN--LLDLVHSDLCEFNGML 498
+I + + I E C C K +K + ++ + +L+L+H DL +
Sbjct: 2119 IIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVE 2298
Query: 499 TRGGNRYFITFIDDCSRYTHVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGEY 558
+ GG RY +DD SR+T V ++ K + F FK ++ + + IK +RSD G E+
Sbjct: 2299 SLGGKRYAYVVVDDFSRFTWVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREF 2478
Query: 559 FSTKFDSFCEEHDIIHECSA 578
+++F FC I HE SA
Sbjct: 2479 ENSRFTEFCTSEGITHEFSA 2538
>BI424202
Length = 421
Score = 75.5 bits (184), Expect(2) = 4e-22
Identities = 38/99 (38%), Positives = 58/99 (58%), Gaps = 1/99 (1%)
Frame = -3
Query: 425 DSVSLWHSRLAHIGISTMNRLIKSKLISC-NIHEFEKCEICVKSKMIKKHFKSVERKFNL 483
+S LWH RL HI I + RL+ ++S + +FE C+K K K K +R NL
Sbjct: 389 ESSMLWHRRLGHISIERIKRLVNEGVLSTLDFADFETYVDCIKGKQTNKSKKGAKRSSNL 210
Query: 484 LDLVHSDLCEFNGMLTRGGNRYFITFIDDCSRYTHVYLL 522
L+++H+D+C + + +YFITFIDD SRY ++Y +
Sbjct: 209 LEIIHTDICCPD--MDANSLKYFITFIDDYSRYMYLYFI 99
Score = 47.8 bits (112), Expect(2) = 4e-22
Identities = 22/39 (56%), Positives = 29/39 (73%)
Frame = -2
Query: 518 HVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGG 556
+V LL K++A + FK +KAEVE Q K IK++RSDRGG
Sbjct: 117 YVSLLHSKNEALDVFKVFKAEVEKQCGKQIKIMRSDRGG 1
>BI784757
Length = 430
Score = 96.7 bits (239), Expect = 3e-20
Identities = 48/119 (40%), Positives = 72/119 (60%), Gaps = 2/119 (1%)
Frame = +1
Query: 459 EKCEICVKSKMIKKHFKSVE--RKFNLLDLVHSDLCEFNGMLTRGGNRYFITFIDDCSRY 516
E C+ C++ K + FK R L++++SD+C + GGNRYFI+FID+ +R
Sbjct: 61 EVCDGCLQCKQSRSTFKQNVPIRAKEKLEVIYSDVCGPMQTESLGGNRYFISFIDELTRK 240
Query: 517 THVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGEYFSTKFDSFCEEHDIIHE 575
VYL++ K D F F+ +K + Q IK+LR++ GGEY ST+F FC++ IIHE
Sbjct: 241 VWVYLIRRKSDFFEVFEKFKNMAKKQSGSLIKILRTNGGGEYVSTEFQEFCDQQGIIHE 417
>TC232910 similar to UP|Q6I8N0 (Q6I8N0) Pol polypeptide, partial (3%)
Length = 690
Score = 87.0 bits (214), Expect = 2e-17
Identities = 55/162 (33%), Positives = 84/162 (50%), Gaps = 5/162 (3%)
Frame = +2
Query: 313 FKTYSESTDDQEVQMRNEVRSKVVGTGTVELNFTSGKKVTLVNVLHVPEMSRDLVSGDLL 372
F + D V M N ++G G V L FTSGK + L +VL VP + ++L+SG +L
Sbjct: 206 FMEFRPIDDGSIVNMGNVATEPILGLGCVNLVFTSGKSLYL-DVLFVPGIRKNLLSGMIL 382
Query: 373 GKPGIKSVYESEKLILTRNGVFVGKGYSAEGMIKL---CPI--DNIINKVSNSAYMIDSV 427
G K V ES+K IL+R+G FVG GY M KL P +++ +S +
Sbjct: 383 NNCGFKQVLESDKYILSRHGSFVGFGYRCNEMFKLNIDVPFVHESVCMASCSSITNMTKS 562
Query: 428 SLWHSRLAHIGISTMNRLIKSKLISCNIHEFEKCEICVKSKM 469
+WH+RL H+ + + K+ +I EKC+ C+ +K+
Sbjct: 563 EIWHARLGHVHYKRLKDMSKTCMIPPFDMNIEKCKTCMLTKI 688
Score = 30.8 bits (68), Expect = 1.7
Identities = 13/25 (52%), Positives = 14/25 (56%)
Frame = +3
Query: 245 KGGCYVCGKPDHFARDCRQNKTKKE 269
K C CGKP H RDCR K K +
Sbjct: 24 KLSCSKCGKPGHLKRDCRVFKGKNK 98
>BI425191 weakly similar to GP|14586969|gb| pol polyprotein {Citrus x
paradisi}, partial (7%)
Length = 423
Score = 83.6 bits (205), Expect = 2e-16
Identities = 44/100 (44%), Positives = 59/100 (59%)
Frame = +3
Query: 458 FEKCEICVKSKMIKKHFKSVERKFNLLDLVHSDLCEFNGMLTRGGNRYFITFIDDCSRYT 517
F ICV K K R LL+++H+D+C + + G RYFITFIDD S Y
Sbjct: 24 FSDLNICVDCIKEKHTMKRATRSTQLLEIMHTDICRPFDVNSFGKERYFITFIDDYSHYG 203
Query: 518 HVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGE 557
+VYLL K A + + + EVE QL++ +KV+RSDRGGE
Sbjct: 204 YVYLLHEKFQAVDVLEIHLNEVERQLDRKVKVVRSDRGGE 323
>TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement pol
polyprotein, partial (4%)
Length = 919
Score = 82.0 bits (201), Expect = 7e-16
Identities = 50/117 (42%), Positives = 65/117 (54%), Gaps = 2/117 (1%)
Frame = +1
Query: 461 CEICVKSKMIKKHFKSVE--RKFNLLDLVHSDLCEFNGMLTRGGNRYFITFIDDCSRYTH 518
C+ C K ++ F + + R LL +VH DLC + T G N YFITFIDD S+
Sbjct: 331 CDTCEIGKKHRESFPTGKSWRMKKLLKIVHLDLCTVE-IPTHGDNNYFITFIDDFSKKMW 507
Query: 519 VYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGEYFSTKFDSFCEEHDIIHE 575
VY LK K +A NAFK +KA E Q +K L D+G EY S + F E+H I H+
Sbjct: 508 VYFLKQKSEACNAFKMFKAFAEKQNGCKVKALIIDKGQEYLS--YTIFFEKHGIQHQ 672
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 77.0 bits (188), Expect = 2e-14
Identities = 68/249 (27%), Positives = 114/249 (45%), Gaps = 4/249 (1%)
Frame = +2
Query: 333 SKVVGTGTVELNFTSGKKV-TLVNVLHVPEMSRDLVSGDLLGKPGIKSVYESEKLILTRN 391
S+VV TG ++ TS + ++V +L P L L +++ ++
Sbjct: 38 SRVVATGIGHVSPTSSLSLNSVVFILGCPFNITSL--SQLTRFRNCSVTFDANSFVIQEC 211
Query: 392 GV--FVGKGYSAEGMIKLCPIDNIINKVSNSAYMIDSVSLWHSRLAHIGISTMNRLIKSK 449
G +G G + G+ L P +S + S L H RL H +S + ++ S
Sbjct: 212 GTGWTIGVGIESHGLYYLKP------NLSWVCSAVTSPKLLHERLGHPHLSKLKIMVPSL 373
Query: 450 LISCNIHEFEKCEICVKSKMIKKHFKSVERKFNLLDLV-HSDLCEFNGMLTRGGNRYFIT 508
++ CE C K ++ + VE + + LV H D+ N + + RYF+T
Sbjct: 374 EKIKDLF----CESCQLGKHVRSSXRHVESRVDSPFLVIHXDIWGPNRVSSMS-YRYFVT 538
Query: 509 FIDDCSRYTHVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGEYFSTKFDSFCE 568
FID+ S+ T V+L+K + + + S +++ Q KTIK+LRSD EYFS+ F
Sbjct: 539 FIDEFSQCTRVFLMKERSEILSFLTSVN-KIKTQFGKTIKILRSDNAKEYFSSVISPFXS 715
Query: 569 EHDIIHECS 577
I+H+ S
Sbjct: 716 AQGILHQFS 742
>BI701169
Length = 407
Score = 72.4 bits (176), Expect = 5e-13
Identities = 34/67 (50%), Positives = 43/67 (63%)
Frame = +2
Query: 506 FITFIDDCSRYTHVYLLKHKDDAFNAFKSYKAEVENQLNKTIKVLRSDRGGEYFSTKFDS 565
F FIDD SR T VY KHK + F FK +KA VE + IK +RSDRGGE+ S +F
Sbjct: 113 FFLFIDDFSRKTWVYFFKHKLEVFENFKKFKAIVEKESGFKIKAMRSDRGGEF*SNEFQK 292
Query: 566 FCEEHDI 572
+C++H I
Sbjct: 293 YCDDHGI 313
>BI321697
Length = 368
Score = 68.2 bits (165), Expect = 1e-11
Identities = 31/87 (35%), Positives = 55/87 (62%)
Frame = -3
Query: 318 ESTDDQEVQMRNEVRSKVVGTGTVELNFTSGKKVTLVNVLHVPEMSRDLVSGDLLGKPGI 377
+ + + V M N + V+G G V+L +SG + L V H+ ++ ++L+S LL + G
Sbjct: 264 QESSTRTVSMGNGSMAHVLGEGQVKLELSSGNFIVLDGVYHISDIRKNLISTSLLVQQGY 85
Query: 378 KSVYESEKLILTRNGVFVGKGYSAEGM 404
K V+ES ++++TR+G F+GKGY +G+
Sbjct: 84 KVVFESNRVVITRHGNFIGKGYICDGL 4
>TC231744
Length = 794
Score = 55.8 bits (133), Expect = 5e-08
Identities = 49/207 (23%), Positives = 82/207 (38%)
Frame = +2
Query: 101 FKAEEEGIKKFLISKYFDFKMLDTKPILQQVHELQVLVNKIKAVKIDIRGAFQVGAIIAK 160
F E+ L+ K K I + + E+ L +K+K++K+++ V ++
Sbjct: 8 FAKNEKAETSNLLDKLISMKYKGKGNIREYIMEISNLASKLKSLKLELGEDLFVHLVLIS 187
Query: 161 LPPSWNGYRKKLLHNYEDFSLEKIQKHLRIEEESKVRDKAESSGFSKANTVTTKGKKKYD 220
LP + ++ + +SL ++ H EEE RD+ ES+ K KK D
Sbjct: 188 LPAHFGQFKVSYNTQKDKWSLNELISHCVQEEERL*RDRTESAH-------NKKRKKTKD 346
Query: 221 GKQQHLGPKKEHNKFKNNNGTKGPKGGCYVCGKPDHFARDCRQNKTKKEVNAVQVDDEII 280
++ KK+ K + CY C K H + C K V+ D +
Sbjct: 347 VAEKTS*QKKQQ---------KDEEFTCYFCKKSRHMKKKC----PKYAAWRVKKDKFLT 487
Query: 281 ATVSEVMAVKGKVPGWWYDTCASVHVS 307
SEV WW D+ A+ H+S
Sbjct: 488 LVCSEVNLAFVPKDTWWVDSGATTHIS 568
>CO984748
Length = 810
Score = 52.8 bits (125), Expect = 4e-07
Identities = 48/203 (23%), Positives = 86/203 (41%), Gaps = 30/203 (14%)
Frame = -2
Query: 264 NKTKKEVNAVQVDDEIIATVSEVMAVKGKVPGWWYDTCASVHVSYDKASFKTYSESTDDQ 323
NK++ + NA Q +A + + W+ + A+ HV+ + ++ ++
Sbjct: 647 NKSQSQQNA-QAPQAYLANANPTQQSQN----WYINYGATHHVTASPQNLMNEVPTSGNE 483
Query: 324 EVQMRNEVRSKVVGTGTVELNFTSGKKVTLVNVLHVPEMSRDLVSGDLLGKPGIKSVYES 383
+V + N + G+ F S K+TL N+LHVP ++++LVS K + + S
Sbjct: 482 QVFLGNGQGLPITGSTVFNSPFASNVKLTLNNLLHVPHITKNLVSVS*FAKDNVFFEFHS 303
Query: 384 EKLI--------------LTRNGVF-------------VGKGYSAEGMIKLCPIDNIINK 416
+ L+++G++ VGK + M L + + N
Sbjct: 302 SHCVVKSQVDGSILLRGALSQDGLYVFSNLLNSSFKFTVGKAAANVHMSSLSDLSSHCNN 123
Query: 417 VSNSAYMIDS---VSLWHSRLAH 436
VS SA + VSLWH+RL H
Sbjct: 122 VSTSALNTSNYSLVSLWHNRLGH 54
>CO981347
Length = 624
Score = 38.9 bits (89), Expect(2) = 9e-07
Identities = 16/31 (51%), Positives = 23/31 (73%)
Frame = +3
Query: 499 TRGGNRYFITFIDDCSRYTHVYLLKHKDDAF 529
T GG+ YF+T IDD SR +Y+LK+K ++F
Sbjct: 18 THGGSSYFLTIIDDFSRRVWLYVLKNKSESF 110
Score = 32.3 bits (72), Expect(2) = 9e-07
Identities = 15/41 (36%), Positives = 23/41 (55%)
Frame = +2
Query: 529 FNAFKSYKAEVENQLNKTIKVLRSDRGGEYFSTKFDSFCEE 569
F F+ + NQL +KVLR+D G E+ +F+ FC +
Sbjct: 107 F*KFRE*HTLIGNQLGTKLKVLRTDNGLEFVLEQFNEFCRK 229
>TC213393 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (23%)
Length = 678
Score = 42.7 bits (99), Expect = 4e-04
Identities = 32/89 (35%), Positives = 49/89 (54%), Gaps = 6/89 (6%)
Frame = -2
Query: 461 CEICVKSKMIKKHFKSV---ERKFNLLDLVHSD---LCEFNGMLTRGGNRYFITFIDDCS 514
CE C K ++ F +F + LVHSD LC +GM T +RYF+TFI+D S
Sbjct: 347 CESCQFGKHVQSSFPYCVICRDRFPFV-LVHSDVWGLC--HGMPTLE-SRYFVTFIEDYS 180
Query: 515 RYTHVYLLKHKDDAFNAFKSYKAEVENQL 543
T ++L+ ++ + F F S+ E++N L
Sbjct: 179 *QTWLFLMVNRSELF*IFTSFYQEIKNNL 93
>CO984791
Length = 672
Score = 42.7 bits (99), Expect = 4e-04
Identities = 35/150 (23%), Positives = 65/150 (43%)
Frame = -3
Query: 112 LISKYFDFKMLDTKPILQQVHELQVLVNKIKAVKIDIRGAFQVGAIIAKLPPSWNGYRKK 171
L++K K I + + E+ L +K+K++K+++ + ++ LP + ++
Sbjct: 607 LLAKLISMKYKGKGNIREYIIEMSNLASKLKSLKLELGEDLLMYLVLISLPAHFGQFKVS 428
Query: 172 LLHNYEDFSLEKIQKHLRIEEESKVRDKAESSGFSKANTVTTKGKKKYDGKQQHLGPKKE 231
+ +SL ++ H EEES RD+ ES+ +T K KK G +K+
Sbjct: 427 YNT*KDKWSLNELISHCVQEEESL*RDRTESAHL--ISTSQNKKWKKTKGAA*GTS*QKK 254
Query: 232 HNKFKNNNGTKGPKGGCYVCGKPDHFARDC 261
N K + CY K H ++C
Sbjct: 253 QN--------KDEEFACYFYKKSGHMKKEC 188
>BF595216
Length = 421
Score = 40.4 bits (93), Expect = 0.002
Identities = 16/39 (41%), Positives = 26/39 (66%)
Frame = +2
Query: 489 SDLCEFNGMLTRGGNRYFITFIDDCSRYTHVYLLKHKDD 527
SDLC ++ G Y++TF+D +RYT ++LL+HK +
Sbjct: 8 SDLCSPAPFVSSTGYSYYVTFVDAFTRYTWIFLLRHKSE 124
>TC226201 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 820
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/38 (47%), Positives = 20/38 (52%), Gaps = 4/38 (10%)
Frame = +1
Query: 229 KKEHNKFKNNNGTKGPKGG----CYVCGKPDHFARDCR 262
K HN G G GG CY CG+P HFAR+CR
Sbjct: 178 KLSHNSRGGGGGRGGRSGGSDLKCYECGEPGHFARECR 291
>BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea mays},
partial (14%)
Length = 421
Score = 38.9 bits (89), Expect = 0.006
Identities = 19/44 (43%), Positives = 25/44 (56%), Gaps = 9/44 (20%)
Frame = -2
Query: 539 VENQLNKTIKVLRSDRGGEYFST---------KFDSFCEEHDII 573
VE Q K IK++RSDRGGEY+ F F +EH+I+
Sbjct: 420 VEKQCGKQIKIVRSDRGGEYYGRYTEDGQAPGSFAKFLQEHEIV 289
>TC226200 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (89%)
Length = 936
Score = 38.5 bits (88), Expect = 0.008
Identities = 17/35 (48%), Positives = 19/35 (53%), Gaps = 4/35 (11%)
Frame = +2
Query: 232 HNKFKNNNGTKGPKGG----CYVCGKPDHFARDCR 262
HN G G GG CY CG+P HFAR+CR
Sbjct: 278 HNSRGGGGGRGGRSGGSDLKCYECGEPGHFARECR 382
>TC226202 similar to UP|O81126 (O81126) 9G8-like SR protein (RSZp22 splicing
factor), partial (72%)
Length = 1003
Score = 38.5 bits (88), Expect = 0.008
Identities = 17/35 (48%), Positives = 19/35 (53%), Gaps = 4/35 (11%)
Frame = +3
Query: 232 HNKFKNNNGTKGPKGG----CYVCGKPDHFARDCR 262
HN G G GG CY CG+P HFAR+CR
Sbjct: 774 HNSRGGGGGRGGRSGGSDLKCYECGEPGHFARECR 878
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,668,040
Number of Sequences: 63676
Number of extensions: 303637
Number of successful extensions: 1804
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 1763
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1797
length of query: 578
length of database: 12,639,632
effective HSP length: 102
effective length of query: 476
effective length of database: 6,144,680
effective search space: 2924867680
effective search space used: 2924867680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0044a.5


