

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0042.1
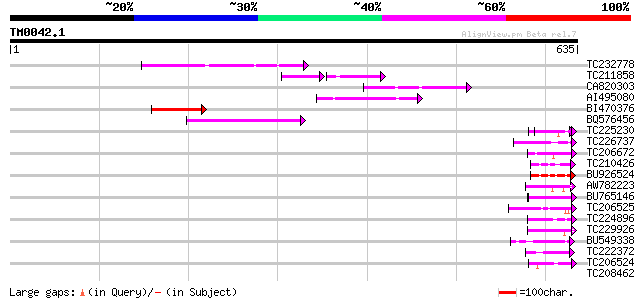
(635 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232778 similar to UP|Q9LNG5 (Q9LNG5) F21D18.16, partial (5%) 70 2e-12
TC211858 43 9e-11
CA820303 65 1e-10
AI495080 62 6e-10
BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sat... 55 7e-08
BQ576456 53 5e-07
TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein ... 46 6e-05
TC226737 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich prot... 45 1e-04
TC206672 weakly similar to UP|Q7UTL3 (Q7UTL3) 3-hydroxyisobutyra... 45 1e-04
TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protei... 44 2e-04
BU926524 similar to GP|9758900|dbj| contains similarity to GAR1 ... 44 2e-04
AW782223 44 3e-04
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 43 5e-04
TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (... 42 6e-04
TC224896 similar to UP|Q94EH8 (Q94EH8) At1g66260/T6J19_1, partia... 42 6e-04
TC229926 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, part... 42 8e-04
BU549338 similar to GP|22531190|gb| transcriptional coactivator-... 42 8e-04
TC222372 similar to UP|Q7KS14 (Q7KS14) CG6695-PB, partial (5%) 42 8e-04
TC206524 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 42 8e-04
TC208462 homologue to UP|LSM4_TOBAC (Q43582) Probable U6 snRNA-a... 42 0.001
>TC232778 similar to UP|Q9LNG5 (Q9LNG5) F21D18.16, partial (5%)
Length = 746
Score = 70.5 bits (171), Expect = 2e-12
Identities = 53/188 (28%), Positives = 82/188 (43%), Gaps = 1/188 (0%)
Frame = -1
Query: 148 GDSDSHRRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMSFGEMTITLD 207
G+ + L+ Q G + + Y +A LI A +ERW ET +FH+ GE TITL
Sbjct: 578 GEKQISEEIVPLLRQCGFYWIMKMGYLKINAALISAFIERWRPETHTFHLRCGEATITLQ 399
Query: 208 DVSALLHLPMGSRFYTPGRGERD-ECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQT 266
DV LL L P G + + A LC +L+G E E + +
Sbjct: 398 DVLILLGLRTDG---APLIGSTNLDWADLCEELLGVRPQEGEIEGSVVKLSWLAHYFSHI 228
Query: 267 RYEAALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVCEYAWGAIA 326
+ E + + R W++ +G +F +KS ++ ++ L D YAWG
Sbjct: 227 NIDEGNVE-QLQRFIRAWILRFIGGVIFVNKSSS-RVSLRYLQFLRDFE*CSTYAWGPAM 54
Query: 327 LATLYDQL 334
LA LY ++
Sbjct: 53 LAYLYREM 30
>TC211858
Length = 524
Score = 43.1 bits (100), Expect(2) = 9e-11
Identities = 21/48 (43%), Positives = 29/48 (59%)
Frame = -1
Query: 305 VYWIGMLEDLGRVCEYAWGAIALATLYDQLSRASRRGTAQMGGFSSLL 352
V ++ DLG+ YAWG AL +Y+QL ASR T Q+ G+ +LL
Sbjct: 464 VVYLDAFRDLGQSGGYAWGVAALVHMYNQLDEASRTTTRQIVGYLTLL 321
Score = 42.0 bits (97), Expect(2) = 9e-11
Identities = 24/70 (34%), Positives = 34/70 (48%), Gaps = 4/70 (5%)
Frame = -3
Query: 355 WVYEYLSD--RVIIRRADPEYSQDQPRARRWAMSRVGHAGLDER--RVMLDELTVDDIIW 410
W+YE+ + +I D Y + PRA RW S+ G+ R D LTV D+ W
Sbjct: 231 WIYEHFPSVHQCVI---DDTY*ETSPRASRWLTSKAHMKGITGAPYRARCDALTVIDVSW 61
Query: 411 TPFEDHRAHR 420
P+ +HR R
Sbjct: 60 LPYTEHRGVR 31
>CA820303
Length = 421
Score = 64.7 bits (156), Expect = 1e-10
Identities = 39/121 (32%), Positives = 60/121 (49%)
Frame = -1
Query: 397 RVMLDELTVDDIIWTPFEDHRAHRPRDPRAMYSRYIRSPFGRVVRRHLPERVLRQFGFIQ 456
R LD L + D+ W P+ +HR + R+ YS + +G VV + ERV+RQFG+ Q
Sbjct: 394 RERLDRLRIPDVCWIPYGEHREVQDFHVRSCYSGLL--CWGPVVVYYRSERVVRQFGYTQ 221
Query: 457 DVPRHPSEIQTSGSLAETADAAFAEFAPHLRPQGIPATYPGEAVEDYMRWYNAVSHRFII 516
+P S + + S + D + + H+ P G PG DY+ W+ SH F+
Sbjct: 220 TIP--ASTVDSWVSYDDIHD-RWMHYEDHIVPAGEVCVVPGACSSDYIDWFFRTSHPFMT 50
Query: 517 P 517
P
Sbjct: 49 P 47
>AI495080
Length = 407
Score = 62.4 bits (150), Expect = 6e-10
Identities = 38/121 (31%), Positives = 61/121 (50%), Gaps = 2/121 (1%)
Frame = +2
Query: 344 QMGGFSSLLLEWVYEYLSDRVIIRRADPEYSQDQPRARRWAMSR--VGHAGLDERRVMLD 401
Q+ G+ +LL W+YE+ V AD +Y + PRA RW ++ V R L+
Sbjct: 5 QLSGYITLLQCWIYEHFPS-VAESTAD*DYDEASPRACRWIATKKTVKSIRTPSYRERLN 181
Query: 402 ELTVDDIIWTPFEDHRAHRPRDPRAMYSRYIRSPFGRVVRRHLPERVLRQFGFIQDVPRH 461
L + D+ W + +HR + R+ YS + +G V + PE+V+RQFG+ +P
Sbjct: 182 RLRISDVCWILYGEHREVQDFHVRSCYSGLLH--WGLVAVYYRPEKVVRQFGYTPAIPAP 355
Query: 462 P 462
P
Sbjct: 356 P 358
>BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 429
Score = 55.5 bits (132), Expect = 7e-08
Identities = 25/62 (40%), Positives = 39/62 (62%)
Frame = -1
Query: 159 LIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMSFGEMTITLDDVSALLHLPMG 218
L++ +GL+ + + + L+ A +ERW ET +FH+ +GE TITL DVS LL +P+
Sbjct: 396 LLQISGLYPIMKLAQLKVNGALVNAFIERWRPETHTFHLKYGEATITLQDVSVLLGIPVD 217
Query: 219 SR 220
R
Sbjct: 216 GR 211
>BQ576456
Length = 424
Score = 52.8 bits (125), Expect = 5e-07
Identities = 41/133 (30%), Positives = 61/133 (45%)
Frame = +1
Query: 199 FGEMTITLDDVSALLHLPMGSRFYTPGRGERDECAALCAQLMGGSVGIYEAEFDTNRSQT 258
+GE+TITLDD++ LLHLP+ + DE L +L+ S A+
Sbjct: 28 WGELTITLDDMACLLHLPITGALHRFEPLGVDEAVLLLMELLEVSREEARAKIVRAHRAY 207
Query: 259 IRFGVLQTRYEAALAEHRYEDAARIWLVNQLGATLFASKSGGYHTTVYWIGMLEDLGRVC 318
+R L+ Y++ + A R +L + + TLFA+KS H V + DL
Sbjct: 208 VRLSWLREVYQSRCQARCWIVAVRAYLFHLVDCTLFANKS-ATHVHVVHLKGF*DLC*SG 384
Query: 319 EYAWGAIALATLY 331
Y WG AL +Y
Sbjct: 385 GYGWGVAALVHMY 423
>TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (66%)
Length = 773
Score = 45.8 bits (107), Expect = 6e-05
Identities = 28/50 (56%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Frame = +1
Query: 588 GRGRGDRARGGRGRGGRARGEGAPAE---GARGGRARGPRGRRGAGRGRG 634
G GRGDR RG RG GGR G P + G RGG RG G RG GRG G
Sbjct: 76 GGGRGDRGRG-RGGGGRGGDRGTPFKARGGGRGGGGRG--GGRGGGRGGG 216
Score = 42.4 bits (98), Expect = 6e-04
Identities = 25/52 (48%), Positives = 27/52 (51%), Gaps = 1/52 (1%)
Frame = +1
Query: 582 GARAAGGRGRGDRARGG-RGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
G R GRGRG RGG RG +ARG G G GGR G G RG +G
Sbjct: 79 GGRGDRGRGRGGGGRGGDRGTPFKARGGGRGGGGRGGGRGGGRGGGRGGMKG 234
Score = 37.0 bits (84), Expect = 0.027
Identities = 22/47 (46%), Positives = 23/47 (48%), Gaps = 6/47 (12%)
Frame = +1
Query: 584 RAAGGRGRGDRAR------GGRGRGGRARGEGAPAEGARGGRARGPR 624
R GGRG GDR GGRG GGR G G G RGG G +
Sbjct: 106 RGGGGRG-GDRGTPFKARGGGRGGGGRGGGRGGGRGGGRGGMKGGSK 243
Score = 33.1 bits (74), Expect = 0.39
Identities = 17/37 (45%), Positives = 18/37 (47%)
Frame = +1
Query: 571 GTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARG 607
G +G F A G R GGRG G G GRGG G
Sbjct: 127 GDRGTPFKARGGGRGGGGRGGGRGGGRGGGRGGMKGG 237
>TC226737 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein, partial
(37%)
Length = 595
Score = 45.1 bits (105), Expect = 1e-04
Identities = 30/70 (42%), Positives = 35/70 (49%)
Frame = -2
Query: 565 RSNAFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPR 624
R++A + + A A G GGRG G R R G +G RARG G R G RG R
Sbjct: 288 RTSAPLRGRAAAAGASGGRGGGGGRGSGARGRTGPRKGARARG------GTRVGGGRGAR 127
Query: 625 GRRGAGRGRG 634
G GAG G G
Sbjct: 126 G--GAGNGTG 103
Score = 38.9 bits (89), Expect = 0.007
Identities = 21/45 (46%), Positives = 23/45 (50%)
Frame = -2
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGR 626
G R +G RGR +G R RGG G G GARGG G GR
Sbjct: 222 GGRGSGARGRTGPRKGARARGGTRVGGG---RGARGGAGNGTGGR 97
>TC206672 weakly similar to UP|Q7UTL3 (Q7UTL3) 3-hydroxyisobutyrate
dehydrogenase , partial (76%)
Length = 1230
Score = 44.7 bits (104), Expect = 1e-04
Identities = 31/61 (50%), Positives = 31/61 (50%), Gaps = 7/61 (11%)
Frame = -3
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARG------EGAPAEGARGGRARG-PRGRRGAGRGR 633
RG R GGR RG R RGG G GR RG RGGR R PRGR GAG R
Sbjct: 358 RGRRRGGGR-RGRRRRGGGGLSGRPRGSRRWRRRRRSGLRRRGGRGRSRPRGRGGAGPWR 182
Query: 634 G 634
G
Sbjct: 181 G 179
>TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protein, partial
(50%)
Length = 434
Score = 43.9 bits (102), Expect = 2e-04
Identities = 28/50 (56%), Positives = 28/50 (56%)
Frame = +3
Query: 584 RAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGR 633
R GG GRG RGGRG GG RG G P RGGR PRG GRGR
Sbjct: 234 RGGGGGGRGG-GRGGRGGGG-FRGRGGP----RGGRGGPPRGGGFRGRGR 365
Score = 36.6 bits (83), Expect = 0.035
Identities = 27/51 (52%), Positives = 28/51 (53%), Gaps = 2/51 (3%)
Frame = +3
Query: 578 AAVRGARAAG-GRGRGDRARGG-RGRGGRARGEGAPAEGARGGRARGPRGR 626
+A RG G G GRG R GG RGRGG G G P RGG RG RGR
Sbjct: 225 SAGRGGGGGGRGGGRGGRGGGGFRGRGGPRGGRGGP---PRGGGFRG-RGR 365
Score = 31.2 bits (69), Expect = 1.5
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Frame = +3
Query: 598 GRGRGGRARGEGAPAEGARGGRARGP-RGRRGAGRGRG 634
GRG GG RG G RGGR G RGR G GRG
Sbjct: 231 GRGGGGGGRG------GGRGGRGGGGFRGRGGPRGGRG 326
>BU926524 similar to GP|9758900|dbj| contains similarity to GAR1
protein~gene_id:MRG7.14 {Arabidopsis thaliana}, partial
(23%)
Length = 341
Score = 43.9 bits (102), Expect = 2e-04
Identities = 30/50 (60%), Positives = 30/50 (60%)
Frame = +2
Query: 584 RAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGR 633
R GG GRG RGGRG GG RG GAP G RGG RG GR G RGR
Sbjct: 80 RGGGGGGRGG-FRGGRGGGG-FRGRGAP-RGGRGGPPRG-GGRGGGFRGR 217
Score = 37.4 bits (85), Expect = 0.021
Identities = 27/52 (51%), Positives = 28/52 (52%), Gaps = 3/52 (5%)
Frame = +2
Query: 578 AAVRGARAAGGRG--RGDRARGG-RGRGGRARGEGAPAEGARGGRARGPRGR 626
+A RG GGRG RG R GG RGRG G G P G GGR G RGR
Sbjct: 71 SAGRGG-GGGGRGGFRGGRGGGGFRGRGAPRGGRGGPPRG--GGRGGGFRGR 217
Score = 29.6 bits (65), Expect = 4.3
Identities = 22/47 (46%), Positives = 22/47 (46%), Gaps = 10/47 (21%)
Frame = +2
Query: 598 GRGRGGRARGEGAPAEGARGGRARG-------PRGRRGA---GRGRG 634
GRG GG RG G RGGR G PRG RG G GRG
Sbjct: 77 GRGGGGGGRG------GFRGGRGGGGFRGRGAPRGGRGGPPRGGGRG 199
>AW782223
Length = 419
Score = 43.5 bits (101), Expect = 3e-04
Identities = 36/73 (49%), Positives = 37/73 (50%), Gaps = 17/73 (23%)
Frame = -2
Query: 578 AAVRGARAAGGRGRGDRA-RGGRGRGGRAR-------GEGAPAEGA--RGG-------RA 620
A R AR +GGRGRG R R RGR GR R G GA EG RGG R
Sbjct: 262 AISRRARRSGGRGRGRRGRRSRRGRRGRRRRPCWTSDGRGARREGGTRRGG*LRRRRSRG 83
Query: 621 RGPRGRRGAGRGR 633
R RGRRG GR R
Sbjct: 82 RSRRGRRG-GRSR 47
Score = 40.4 bits (93), Expect = 0.002
Identities = 35/91 (38%), Positives = 40/91 (43%), Gaps = 12/91 (13%)
Frame = -2
Query: 549 EGTHSRSLTERALDLIRSNAFIGTQGVAFAAVRGARAAGGR---------GRGDRARGGR 599
E T SR+++ RA RS G +G R R GR GRG R GG
Sbjct: 280 E*TESRAISRRAR---RS----GGRGRGRRGRRSRRGRRGRRRRPCWTSDGRGARREGGT 122
Query: 600 GRGG---RARGEGAPAEGARGGRARGPRGRR 627
RGG R R G G RGGR+R GRR
Sbjct: 121 RRGG*LRRRRSRGRSRRGRRGGRSRSSGGRR 29
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 42.7 bits (99), Expect = 5e-04
Identities = 23/53 (43%), Positives = 24/53 (44%)
Frame = +2
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G + GG G G GG G GG RGE G RGG G G G G G G
Sbjct: 149 GGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGG 307
Score = 42.0 bits (97), Expect = 8e-04
Identities = 23/54 (42%), Positives = 24/54 (43%)
Frame = +2
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
RG GG G G GG+G GG G G G GG RG R GRG G
Sbjct: 101 RGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGG 262
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/53 (41%), Positives = 23/53 (42%)
Frame = +2
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G GG G G GG GRGG G G G GG+ G G G G G G
Sbjct: 44 GGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGGG 202
Score = 40.0 bits (92), Expect = 0.003
Identities = 22/53 (41%), Positives = 23/53 (42%)
Frame = +2
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G GG RG+R G GRGG G G G GG G G G G G G
Sbjct: 194 GGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGG 352
Score = 37.4 bits (85), Expect = 0.021
Identities = 24/67 (35%), Positives = 26/67 (37%)
Frame = +3
Query: 569 FIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRG 628
F+G + G GG G G G G GGR G G G GG G G G
Sbjct: 216 FLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGGGRGEGGGG---GGGGGGGGGGGGEGG 386
Query: 629 AGRGRGE 635
RG GE
Sbjct: 387 GRRGGGE 407
Score = 37.4 bits (85), Expect = 0.021
Identities = 22/53 (41%), Positives = 23/53 (42%), Gaps = 2/53 (3%)
Frame = +2
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGP--RGRRGAGRG 632
G GG G G GG G GG+ G G G GG G RG R GRG
Sbjct: 89 GGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRG 247
Score = 35.0 bits (79), Expect = 0.10
Identities = 24/58 (41%), Positives = 25/58 (42%), Gaps = 5/58 (8%)
Frame = +1
Query: 582 GARAAGGRG---RGDRARGG--RGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
G GG G G+R R G RG GG G G G GG G RG G G G G
Sbjct: 187 GGGGGGGGGVF*GGERXREGGERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGGGGG 360
>TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (clone
na-481-5), partial (20%)
Length = 752
Score = 42.4 bits (98), Expect = 6e-04
Identities = 38/92 (41%), Positives = 44/92 (47%), Gaps = 16/92 (17%)
Frame = +2
Query: 559 RALDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGG 618
+AL+L + T V A R + +GGRG G R+ GG GR G ARG G GARGG
Sbjct: 197 KALELHETELGGYTLTVDEAKPRDNQGSGGRGAGGRS-GGGGRFG-ARGGGGGRFGARGG 370
Query: 619 RAR------GPR----------GRRGAGRGRG 634
R G R GR G GRGRG
Sbjct: 371 GGRFGGSGGGGRFGGSGGGRFGGRGGGGRGRG 466
Score = 38.9 bits (89), Expect = 0.007
Identities = 28/60 (46%), Positives = 29/60 (47%), Gaps = 6/60 (10%)
Frame = +2
Query: 582 GARAAGGRGRGDRARGGR----GRGGRARGEGAPAEGARGGRARGPRGRRG--AGRGRGE 635
GAR GG G R GGR G GGR G G G RGG RG RG R A G G+
Sbjct: 326 GARGGGGGRFGARGGGGRFGGSGGGGRFGGSGGGRFGGRGGGGRG-RGNRPNLAAEGTGK 502
>TC224896 similar to UP|Q94EH8 (Q94EH8) At1g66260/T6J19_1, partial (28%)
Length = 1102
Score = 42.4 bits (98), Expect = 6e-04
Identities = 28/55 (50%), Positives = 29/55 (51%), Gaps = 1/55 (1%)
Frame = +2
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGA-GRGRG 634
RG +A GG R G RGG G G G GGR RG RGR GA GRGRG
Sbjct: 209 RGGQAGGGPAMPTRGAGWGRRGGPRSGSG---RGGAGGRGRG-RGRGGAGGRGRG 361
Score = 35.8 bits (81), Expect = 0.060
Identities = 22/44 (50%), Positives = 24/44 (54%), Gaps = 4/44 (9%)
Frame = +2
Query: 595 ARGGRGRGGRA---RGEG-APAEGARGGRARGPRGRRGAGRGRG 634
+RGG+ GG A RG G G R G RG G RG GRGRG
Sbjct: 206 SRGGQAGGGPAMPTRGAGWGRRGGPRSGSGRGGAGGRGRGRGRG 337
Score = 32.7 bits (73), Expect = 0.51
Identities = 20/51 (39%), Positives = 23/51 (44%)
Frame = +2
Query: 572 TQGVAFAAVRGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARG 622
T+G + G R+ GRG G GGR RG G G GGR RG
Sbjct: 245 TRGAGWGRRGGPRSGSGRG---------GAGGRGRGRG---RGGAGGRGRG 361
Score = 30.0 bits (66), Expect = 3.3
Identities = 15/33 (45%), Positives = 17/33 (51%)
Frame = +2
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAE 613
RG GRGRG GGRGRG + E + E
Sbjct: 296 RGGAGGRGRGRGRGGAGGRGRGRKDAVEKSAEE 394
>TC229926 similar to UP|Q93VB2 (Q93VB2) AT3g62770/F26K9_200, partial (46%)
Length = 829
Score = 42.0 bits (97), Expect = 8e-04
Identities = 27/59 (45%), Positives = 29/59 (48%), Gaps = 5/59 (8%)
Frame = -2
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGR-----ARGPRGRRGAGRGRG 634
RG + GRG R R GR RGGR+RG G GR R P RRG G GRG
Sbjct: 411 RGCCSGRACGRGRRRRRGRSRGGRSRGRGRSCRCGSRGRWRRRSTRSPD*RRGGG-GRG 238
Score = 34.7 bits (78), Expect = 0.13
Identities = 25/61 (40%), Positives = 26/61 (41%), Gaps = 8/61 (13%)
Frame = -2
Query: 582 GARAAGGRGRGDRAR--------GGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGR 633
G RA G G R R GGRGR R R G + A G R RGR GR R
Sbjct: 513 GRRARRCSGTGGRPRS*PCWEGSGGRGRSRRRRRRGCCSGRACGRGRRRRRGRSRGGRSR 334
Query: 634 G 634
G
Sbjct: 333 G 331
Score = 29.3 bits (64), Expect = 5.6
Identities = 21/51 (41%), Positives = 22/51 (42%), Gaps = 1/51 (1%)
Frame = -2
Query: 584 RAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARG-PRGRRGAGRGR 633
R + GRGR RGG GR G A GGR R P GRGR
Sbjct: 582 RRSCGRGRRRFGRGGGGRP*PPSGRRARRCSGTGGRPRS*PCWEGSGGRGR 430
>BU549338 similar to GP|22531190|gb| transcriptional coactivator-like protein
{Arabidopsis thaliana}, partial (22%)
Length = 565
Score = 42.0 bits (97), Expect = 8e-04
Identities = 31/73 (42%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Frame = -3
Query: 561 LDLIRSNAFIGTQGVAFAAVRGARAAGGRGRGDRA-RGGRGRGGRARGEGAPAEGARGGR 619
++++ +N I T GVA A G G D R G+GRGG R G +G R R
Sbjct: 548 IEIVGTN--ISTPGVAPAR------NGAIGNXDGVPRSGQGRGGALRRPGGRGQGVRRDR 393
Query: 620 ARGPRGRRGAGRG 632
RG RGR GAGRG
Sbjct: 392 GRG-RGRGGAGRG 357
Score = 33.5 bits (75), Expect = 0.30
Identities = 19/33 (57%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Frame = -3
Query: 584 RAAGGRGRG---DRARGGRGRGGRARGEGAPAE 613
R GGRG+G DR RG RGRGG RGE A+
Sbjct: 434 RRPGGRGQGVRRDRGRG-RGRGGAGRGEKVSAD 339
>TC222372 similar to UP|Q7KS14 (Q7KS14) CG6695-PB, partial (5%)
Length = 465
Score = 42.0 bits (97), Expect = 8e-04
Identities = 28/56 (50%), Positives = 30/56 (53%), Gaps = 1/56 (1%)
Frame = -2
Query: 578 AAVRG-ARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRG 632
A +RG A AAGGRG GG GR ARG P +GAR G G RGAG G
Sbjct: 155 APLRGRAAAAGGRG------GGGGRRYGARGRTGPRKGARARGGTGVGGGRGAGNG 6
>TC206524 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 464
Score = 42.0 bits (97), Expect = 8e-04
Identities = 29/61 (47%), Positives = 31/61 (50%), Gaps = 8/61 (13%)
Frame = +3
Query: 582 GARAAGGR------GRGDRARGGRGRGGRARGEGAPAEGARGGRARGPR--GRRGAGRGR 633
G R+ GGR G G GGRG GGR G G G R G + G R GR G GRGR
Sbjct: 33 GGRSGGGRFGGRSGGGGGGRFGGRGGGGRFGGSGG---GGRFGGSGGGRFGGRGGGGRGR 203
Query: 634 G 634
G
Sbjct: 204 G 206
Score = 38.9 bits (89), Expect = 0.007
Identities = 27/56 (48%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Frame = +3
Query: 582 GARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRG--AGRGRGE 635
G GGRG G R GG G GGR G G G RGG RG RG R A G G+
Sbjct: 81 GGGRFGGRGGGGRF-GGSGGGGRFGGSGGGRFGGRGGGGRG-RGNRPNLAAEGTGK 242
Score = 33.5 bits (75), Expect = 0.30
Identities = 19/42 (45%), Positives = 23/42 (54%)
Frame = +3
Query: 593 DRARGGRGRGGRARGEGAPAEGARGGRARGPRGRRGAGRGRG 634
++ GGRG GGR+ G G GGR+ G G R GRG G
Sbjct: 6 NQGSGGRGAGGRSGG------GRFGGRSGGGGGGRFGGRGGG 113
>TC208462 homologue to UP|LSM4_TOBAC (Q43582) Probable U6 snRNA-associated
Sm-like protein LSm4 (Glycine-rich protein 10) (GRP 10),
partial (94%)
Length = 794
Score = 41.6 bits (96), Expect = 0.001
Identities = 27/59 (45%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Frame = +3
Query: 582 GARAAGGRGRGDRARGGRGRG-GRARGEGAPAEGARGGRARG-----PRGRRGAGRGRG 634
G GRGR D G + +G GR EG P +GGR RG P G RG GRGRG
Sbjct: 405 GVGRGRGRGREDGPGGRQPKGIGRGLDEGGPK--GQGGRGRGGPGGKPGGNRGGGRGRG 575
Score = 30.4 bits (67), Expect = 2.5
Identities = 17/42 (40%), Positives = 19/42 (44%)
Frame = +3
Query: 581 RGARAAGGRGRGDRARGGRGRGGRARGEGAPAEGARGGRARG 622
RG G +G+G R RGG G G P GGR RG
Sbjct: 474 RGLDEGGPKGQGGRGRGGPG--------GKPGGNRGGGRGRG 575
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,480,134
Number of Sequences: 63676
Number of extensions: 394241
Number of successful extensions: 5441
Number of sequences better than 10.0: 434
Number of HSP's better than 10.0 without gapping: 4109
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4954
length of query: 635
length of database: 12,639,632
effective HSP length: 103
effective length of query: 532
effective length of database: 6,081,004
effective search space: 3235094128
effective search space used: 3235094128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0042.1


