

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.4
(266 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
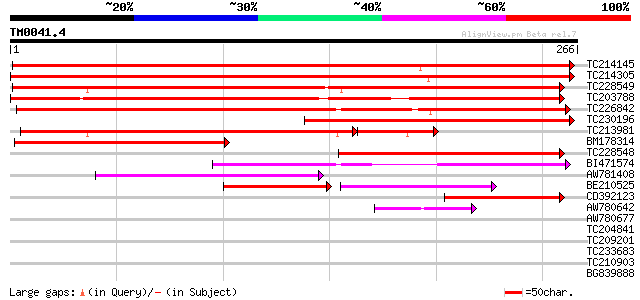
Score E
Sequences producing significant alignments: (bits) Value
TC214145 similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated pro... 456 e-129
TC214305 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associa... 441 e-124
TC228549 similar to GB|AAP13420.1|30023774|BT006312 At3g45600 {A... 271 2e-73
TC203788 similar to GB|AAL91270.1|19699320|AY090367 AT3g12090/T2... 246 8e-66
TC226842 weakly similar to UP|Q84VG2 (Q84VG2) Senescence-associa... 244 2e-65
TC230196 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associa... 236 1e-62
TC213981 similar to GB|AAP13420.1|30023774|BT006312 At3g45600 {A... 152 5e-43
BM178314 weakly similar to PIR|T02906|T029 senescence-associated... 163 7e-41
TC228548 similar to GB|AAP13420.1|30023774|BT006312 At3g45600 {A... 129 2e-30
BI471574 127 4e-30
AW781408 similar to GP|19699320|gb| AT3g12090/T21B14_110 {Arabid... 94 6e-20
BE210525 60 1e-16
CD392123 62 3e-10
AW780642 46 2e-05
AW780677 35 0.035
TC204841 similar to UP|Q9LQL8 (Q9LQL8) F5D14.17 protein, partial... 33 0.17
TC209201 similar to UP|Q8H0S2 (Q8H0S2) At4g23490/F16G20_190, par... 32 0.39
TC233683 31 0.66
TC210903 25 0.88
BG839888 29 1.9
>TC214145 similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated protein-like,
partial (87%)
Length = 1867
Score = 456 bits (1174), Expect = e-129
Identities = 209/265 (78%), Positives = 238/265 (88%), Gaps = 1/265 (0%)
Frame = +1
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGL 61
VR SNN+IGLLNFLT LLSIPILV GVWLSKQ T+CERWLE+P+IALGVFL+LVSLAGL
Sbjct: 862 VRFSNNVIGLLNFLTFLLSIPILVAGVWLSKQGATECERWLEKPVIALGVFLMLVSLAGL 1041
Query: 62 VGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNW 121
VGACCRVSWLLW+YLLVMF+LIVL+FAFTIFAF VTNKGAGE +S RGY+EYRLGDYSNW
Sbjct: 1042 VGACCRVSWLLWLYLLVMFVLIVLLFAFTIFAFVVTNKGAGEVVSNRGYKEYRLGDYSNW 1221
Query: 122 LQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGF 181
LQKRVN+T WNRIRSCL SGK+C+EF+++F+NDT +FY+ENLSALQSGCCKP+ +C F
Sbjct: 1222 LQKRVNNTKTWNRIRSCLQSGKVCTEFQSKFLNDTVTEFYSENLSALQSGCCKPAEECQF 1401
Query: 182 TYQGPSVWNK-TEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVI 240
+Y P+ W K T N SNPDC AW+NDP VLCFNCQSCKAGLLQN+KTDWK+VAVVN++
Sbjct: 1402 SYVNPTTWTKPTNVTNQSNPDCDAWNNDPTVLCFNCQSCKAGLLQNLKTDWKRVAVVNIV 1581
Query: 241 FLVFLIIVYSIGCCAFRNNRNDNWK 265
FLVFLIIVYSIGCCAFRNNR +NWK
Sbjct: 1582 FLVFLIIVYSIGCCAFRNNRRENWK 1656
>TC214305 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated
protein-like, partial (89%)
Length = 1003
Score = 441 bits (1133), Expect = e-124
Identities = 202/266 (75%), Positives = 234/266 (87%), Gaps = 1/266 (0%)
Frame = +1
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
MVR SNN+IGLLNFLT LLSIPILV GVWLSKQ T+CERWLE+P+IALGVFL+LVSLAG
Sbjct: 67 MVRFSNNVIGLLNFLTFLLSIPILVAGVWLSKQGATECERWLEKPVIALGVFLMLVSLAG 246
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
LVGACCRVSWLLW+YLLVMF+LIVL+FAFTIFAF VTNKGAGE +S RGY+EYRLGDYSN
Sbjct: 247 LVGACCRVSWLLWLYLLVMFVLIVLLFAFTIFAFVVTNKGAGEVVSNRGYKEYRLGDYSN 426
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
WLQK+VN+T WNRI SCL SGK+C+EF+++F+NDT QFY E+LSALQSGCCKP+ +C
Sbjct: 427 WLQKKVNNTKTWNRISSCLHSGKVCTEFQSKFLNDTVTQFYTEHLSALQSGCCKPAEECL 606
Query: 181 FTYQGPSVWNKTEGV-NYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNV 239
FTY+ + W K V +Y+NPDC AW+N+ VLCFNCQSCKAG LQN KT+WK+VAVVN+
Sbjct: 607 FTYENSTSWTKPGNVTSYNNPDCDAWNNNQTVLCFNCQSCKAGFLQNFKTEWKRVAVVNI 786
Query: 240 IFLVFLIIVYSIGCCAFRNNRNDNWK 265
+FLV LIIVYSIGCCAFRNNR +NWK
Sbjct: 787 VFLVLLIIVYSIGCCAFRNNRRENWK 864
>TC228549 similar to GB|AAP13420.1|30023774|BT006312 At3g45600 {Arabidopsis
thaliana;} , complete
Length = 1010
Score = 271 bits (693), Expect = 2e-73
Identities = 129/263 (49%), Positives = 179/263 (68%), Gaps = 4/263 (1%)
Frame = +2
Query: 2 VRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPG-TDCERWLERPIIALGVFLLLVSLAG 60
+R SN+LIGLLNFLT LLSIPIL G+WLS + TDC ++L+ P+I +GV +++VSLAG
Sbjct: 11 MRTSNHLIGLLNFLTFLLSIPILGGGIWLSSRANNTDCLKFLQWPLIIIGVSIMVVSLAG 190
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
GAC R ++L+ +YL+VMFL+I ++ F IFA+ VT+KG+G + R Y EY L DYS
Sbjct: 191 FAGACYRNTFLMRLYLVVMFLVIAVLIGFIIFAYVVTDKGSGRRVMNRAYLEYYLEDYSG 370
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVN---DTAEQFYAENLSALQSGCCKPSN 177
WL++RV S W +I SC+ K+C R VN T + FY +L+ +QSGCCKP
Sbjct: 371 WLEERVASESYWGKIASCIRDSKVCGRM-GRTVNGMPQTPDMFYLTHLTPIQSGCCKPPT 547
Query: 178 DCGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVV 237
DCG+ YQ +VW G+ +N DC W ND LC+ C SCKAG+L ++K W+KV+V+
Sbjct: 548 DCGYVYQNETVWIPGSGLMGTNADCTRWSNDQEQLCYACDSCKAGVLASLKKSWRKVSVI 727
Query: 238 NVIFLVFLIIVYSIGCCAFRNNR 260
N++ ++ L+IVY I A+RNNR
Sbjct: 728 NIVVMIILVIVYIIAYAAYRNNR 796
>TC203788 similar to GB|AAL91270.1|19699320|AY090367 AT3g12090/T21B14_110
{Arabidopsis thaliana;} , partial (84%)
Length = 1321
Score = 246 bits (628), Expect = 8e-66
Identities = 117/260 (45%), Positives = 163/260 (62%)
Frame = +1
Query: 1 MVRLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAG 60
M R SN +IG LN TLL SIPI+ G+W+++ T CE +L+ P++ +G +L+VSLAG
Sbjct: 163 MYRFSNTVIGFLNLFTLLASIPIIGAGLWMARS-STTCENFLQTPLLVIGFVVLVVSLAG 339
Query: 61 LVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSN 120
+GAC V+ LW+YL+VM LI + TIF F VT+KG G + GR Y+EY L DYS
Sbjct: 340 FIGACFHVACALWLYLVVMLFLIAALMGLTIFGFGVTSKGGGVEVPGRVYKEYHLQDYSP 519
Query: 121 WLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCG 180
WL+KR+ WN IR C+ K C + + T + ++S +QSGCCKP C
Sbjct: 520 WLRKRIQDPRYWNTIRGCILGSKTC----EKLASWTPLDYMQRDMSPIQSGCCKPPTAC- 684
Query: 181 FTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVI 240
+N + +PDC+ W+N PN+LC+ C SCKAG+L++I+ +W K++V+ V
Sbjct: 685 -------TYNVATTMMTQDPDCYRWNNAPNLLCYECDSCKAGVLEDIRGNWHKLSVLTVT 843
Query: 241 FLVFLIIVYSIGCCAFRNNR 260
LV LI +YSIGCCAFRN R
Sbjct: 844 MLVLLIGIYSIGCCAFRNTR 903
>TC226842 weakly similar to UP|Q84VG2 (Q84VG2) Senescence-associated
protein-like protein, partial (30%)
Length = 1116
Score = 244 bits (624), Expect = 2e-65
Identities = 114/261 (43%), Positives = 167/261 (63%), Gaps = 1/261 (0%)
Frame = +1
Query: 4 LSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLVG 63
+SNN+ +LN + L SIPI+ +G+WL+ +P +C PI+ +G+ +LLVSLAG VG
Sbjct: 19 VSNNITAVLNLIATLASIPIIASGIWLASKPDNECVANFRWPIVIIGILILLVSLAGFVG 198
Query: 64 ACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQ 123
A LL +YL M LLI L+ +FAF VT + GRGY+EYRL +S+WL+
Sbjct: 199 AYWNKQGLLALYLFCMALLIALLLLVLVFAFVVTRPDGAYDVPGRGYKEYRLHGFSSWLR 378
Query: 124 KRVNSTGNWNRIRSCLGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTY 183
V +G+W +IR CL + +CS+ ++ TA+QF+ ++S LQSGCCKP CG+ Y
Sbjct: 379 NHVTGSGSWQKIRPCLAASDVCSKLTQDYI--TADQFFNSHISPLQSGCCKPPTACGYNY 552
Query: 184 QGPSVWNKTEGVN-YSNPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFL 242
P +W T VN ++ DC+ W+ND N LC+NC +CKAGLL N++ +W+K ++ ++ +
Sbjct: 553 VNPILW--TNPVNPMADSDCYLWNNDQNQLCYNCNACKAGLLGNLRKEWRKANIILIVAV 726
Query: 243 VFLIIVYSIGCCAFRNNRNDN 263
V LI VY I C AFRN + +N
Sbjct: 727 VVLIWVYVIACSAFRNAQTEN 789
>TC230196 weakly similar to UP|Q9SUD4 (Q9SUD4) Senescence-associated
protein-like, partial (37%)
Length = 662
Score = 236 bits (601), Expect = 1e-62
Identities = 102/127 (80%), Positives = 113/127 (88%)
Frame = +2
Query: 139 LGSGKLCSEFRNRFVNDTAEQFYAENLSALQSGCCKPSNDCGFTYQGPSVWNKTEGVNYS 198
L S KLC +F +F ND+A+QFYAENLSALQSGCCKPSNDC F YQGPSVWNKT+GVN+S
Sbjct: 8 LQSAKLCDKFETQFANDSAQQFYAENLSALQSGCCKPSNDCNFAYQGPSVWNKTDGVNHS 187
Query: 199 NPDCHAWDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLVFLIIVYSIGCCAFRN 258
NPDC+AWDND NVLCFNC+SCKAG LQN+KTDWKKV +VNVIFLVFLIIVYS+GCCAFRN
Sbjct: 188 NPDCNAWDNDSNVLCFNCESCKAGFLQNLKTDWKKVTIVNVIFLVFLIIVYSVGCCAFRN 367
Query: 259 NRNDNWK 265
N NWK
Sbjct: 368 NMRGNWK 388
>TC213981 similar to GB|AAP13420.1|30023774|BT006312 At3g45600 {Arabidopsis
thaliana;} , partial (62%)
Length = 625
Score = 152 bits (384), Expect(2) = 5e-43
Identities = 77/161 (47%), Positives = 109/161 (66%), Gaps = 3/161 (1%)
Frame = +1
Query: 6 NNLIGLLNFLTLLLSIPILVTGVWLSKQPG-TDCERWLERPIIALGVFLLLVSLAGLVGA 64
N+LIGLLNFLT LLSIPIL G+WLS + TDC ++L+ P+I +GV +++VSLAG GA
Sbjct: 1 NHLIGLLNFLTFLLSIPILGGGIWLSSRANNTDCLKFLQWPLIIIGVSIMVVSLAGFAGA 180
Query: 65 CCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGEALSGRGYREYRLGDYSNWLQK 124
C R ++L+ +YL+VMFL+I ++ F IFA+ VT+KG+G + R Y EY L DYS WL++
Sbjct: 181 CYRNTFLMRLYLVVMFLVIAVLIGFIIFAYVVTDKGSGRRVMNRAYLEYYLEDYSGWLEE 360
Query: 125 RVNSTGNWNRIRSCLGSGKLCSEFRNRF--VNDTAEQFYAE 163
RV S W +I SC+ K C + +T++ FY +
Sbjct: 361 RVASDSYWGKIVSCVRDSKACGRMGITINGMPETSDMFYXK 483
Score = 39.7 bits (91), Expect(2) = 5e-43
Identities = 18/39 (46%), Positives = 25/39 (63%), Gaps = 1/39 (2%)
Frame = +2
Query: 164 NLSALQSGCCKPSNDCGFTYQG-PSVWNKTEGVNYSNPD 201
+L+ +QS CCKP DCG+ YQ SV + +G +NPD
Sbjct: 485 HLTPIQSXCCKPPTDCGYVYQNETSVDPQDQGXMGANPD 601
>BM178314 weakly similar to PIR|T02906|T029 senescence-associated protein
homolog T13J8.160 - Arabidopsis thaliana, partial (37%)
Length = 426
Score = 163 bits (413), Expect = 7e-41
Identities = 77/101 (76%), Positives = 91/101 (89%)
Frame = +1
Query: 3 RLSNNLIGLLNFLTLLLSIPILVTGVWLSKQPGTDCERWLERPIIALGVFLLLVSLAGLV 62
RLSN+L+GLLN L+ LLSIPILVTGVWL KQ T+CERWLE+P+I LGVFLL+VSLAGL+
Sbjct: 124 RLSNSLVGLLNLLSFLLSIPILVTGVWLHKQAETECERWLEKPLIVLGVFLLVVSLAGLL 303
Query: 63 GACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKGAGE 103
GACCR+S LLW+YL VMF+LI++V AFT+FAF VTNKGAGE
Sbjct: 304 GACCRLSCLLWLYLFVMFILIIVVSAFTVFAFVVTNKGAGE 426
>TC228548 similar to GB|AAP13420.1|30023774|BT006312 At3g45600 {Arabidopsis
thaliana;} , partial (46%)
Length = 690
Score = 129 bits (323), Expect = 2e-30
Identities = 51/106 (48%), Positives = 73/106 (68%)
Frame = +1
Query: 155 DTAEQFYAENLSALQSGCCKPSNDCGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCF 214
+T + FY +L+ +QSGCCKP DCG+ YQ +VW G+ +NPDC W ND LC+
Sbjct: 4 ETPDMFYIRHLTPIQSGCCKPPTDCGYVYQNETVWIPGSGLMGANPDCTRWSNDQEQLCY 183
Query: 215 NCQSCKAGLLQNIKTDWKKVAVVNVIFLVFLIIVYSIGCCAFRNNR 260
C SCKAG+L ++K W+KV+V+N++ ++ L+IVY I A+RNNR
Sbjct: 184 ACDSCKAGVLASLKKSWRKVSVINIVVMIILVIVYIIAYAAYRNNR 321
>BI471574
Length = 422
Score = 127 bits (320), Expect = 4e-30
Identities = 61/168 (36%), Positives = 93/168 (55%)
Frame = +1
Query: 96 VTNKGAGEALSGRGYREYRLGDYSNWLQKRVNSTGNWNRIRSCLGSGKLCSEFRNRFVND 155
VT + GRGY+EYRL +S+WL+ V +G+W +IR CL + +CS+ ++
Sbjct: 1 VTRPDGAYDVPGRGYKEYRLHGFSSWLRNHVTGSGSWQKIRPCLAASDVCSKLTQDYI-- 174
Query: 156 TAEQFYAENLSALQSGCCKPSNDCGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFN 215
TA+QF+ ++S LQS DC+ W+ND N LC+N
Sbjct: 175 TADQFFNSHISPLQS------------------------------DCYLWNNDQNQLCYN 264
Query: 216 CQSCKAGLLQNIKTDWKKVAVVNVIFLVFLIIVYSIGCCAFRNNRNDN 263
C +CKAGLL N++ +W+K ++ ++ +V LI VY I C AFRN + +N
Sbjct: 265 CNACKAGLLGNLRKEWRKANIILIVAVVVLIWVYVIACSAFRNAQTEN 408
>AW781408 similar to GP|19699320|gb| AT3g12090/T21B14_110 {Arabidopsis
thaliana}, partial (23%)
Length = 367
Score = 94.0 bits (232), Expect = 6e-20
Identities = 38/107 (35%), Positives = 64/107 (59%)
Frame = +2
Query: 41 WLERPIIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAFAVTNKG 100
+ + P++ +G +L++SLAG +GAC V+W LWVYL++M +I + F +F F VT +G
Sbjct: 5 FFQTPLLVIGFVVLVISLAGFIGACFHVAWALWVYLVIMVFIIAALLWFAVFGFVVTEQG 184
Query: 101 AGEALSGRGYREYRLGDYSNWLQKRVNSTGNWNRIRSCLGSGKLCSE 147
G + + ++ Y L YS WL+ R+ W+ I SC+ C++
Sbjct: 185 GGVEVPVKAFK*YCLDRYSPWLRNRIKDPQYWSSIMSCIWGSNTCAK 325
>BE210525
Length = 381
Score = 60.1 bits (144), Expect(2) = 1e-16
Identities = 25/73 (34%), Positives = 42/73 (57%)
Frame = +1
Query: 156 TAEQFYAENLSALQSGCCKPSNDCGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFN 215
T +Q+ + LS +++GCC+P + CG+ S ++ T N DC + N + C++
Sbjct: 163 TLKQYKSAKLSPIEAGCCRPPSQCGYPAVNASYYDLTFHPVSPNNDCKRYKNSRAIKCYD 342
Query: 216 CQSCKAGLLQNIK 228
C SCKAG+ Q +K
Sbjct: 343 CDSCKAGVAQYMK 381
Score = 43.1 bits (100), Expect(2) = 1e-16
Identities = 16/51 (31%), Positives = 32/51 (62%)
Frame = +3
Query: 101 AGEALSGRGYREYRLGDYSNWLQKRVNSTGNWNRIRSCLGSGKLCSEFRNR 151
+G +++G Y+EY+L D+S+ K +N++ NW R++ CL + C+ +
Sbjct: 3 SGHSVTGLRYKEYQLQDFSSLFLKELNNSRNWERLKVCLVKSEDCNNLSKK 155
>CD392123
Length = 601
Score = 62.0 bits (149), Expect = 3e-10
Identities = 28/56 (50%), Positives = 37/56 (66%)
Frame = -2
Query: 205 WDNDPNVLCFNCQSCKAGLLQNIKTDWKKVAVVNVIFLVFLIIVYSIGCCAFRNNR 260
W P SCKAG+L++I+ +W K++V+ V LV LI +YSIGCCAFRN R
Sbjct: 417 WIKTPRAEFGTRDSCKAGVLEDIRGNWHKLSVLTVTMLVLLIGIYSIGCCAFRNTR 250
>AW780642
Length = 142
Score = 45.8 bits (107), Expect = 2e-05
Identities = 17/48 (35%), Positives = 26/48 (53%)
Frame = +2
Query: 172 CCKPSNDCGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSC 219
CCKP CG+ Y P +W + ++ C+ W ND N LC++ +C
Sbjct: 2 CCKPPTACGYYYVNPILWTNPV-IPMADSYCYLWSNDLNHLCYHSDAC 142
>AW780677
Length = 166
Score = 35.0 bits (79), Expect = 0.035
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Frame = +2
Query: 166 SALQSGCCKPSNDCGFTYQGPSVWNKTEGVN-YSNPDCHAWDNDPNVLCFN--CQSC 219
S LQS C KP+ CG+ P +W T VN ++ D + W+ D N L + C C
Sbjct: 2 SPLQSRCSKPTTACGYI*FNPILW--TNPVNPMADSDVYLWNIDHNQLFYT**CVQC 166
>TC204841 similar to UP|Q9LQL8 (Q9LQL8) F5D14.17 protein, partial (91%)
Length = 1302
Score = 32.7 bits (73), Expect = 0.17
Identities = 18/54 (33%), Positives = 26/54 (47%)
Frame = +1
Query: 41 WLERPIIALGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLLIVLVFAFTIFAF 94
W I +GV L L+S G +GA R L Y +++ LLI++ F F
Sbjct: 361 WFIYLFIGIGVVLFLISCFGCIGAATRNGCCLSCYSILVALLILVELGCAAFIF 522
>TC209201 similar to UP|Q8H0S2 (Q8H0S2) At4g23490/F16G20_190, partial (53%)
Length = 993
Score = 31.6 bits (70), Expect = 0.39
Identities = 15/46 (32%), Positives = 20/46 (42%)
Frame = +1
Query: 175 PSNDCGFTYQGPSVWNKTEGVNYSNPDCHAWDNDPNVLCFNCQSCK 220
P DC + P+ ++K E Y PD H WD P C + K
Sbjct: 751 PHPDCRWKMANPAAFDKVEV--YKKPDPHLWDRAPRRNCCRVRKSK 882
>TC233683
Length = 624
Score = 30.8 bits (68), Expect = 0.66
Identities = 16/34 (47%), Positives = 22/34 (64%)
Frame = +1
Query: 49 LGVFLLLVSLAGLVGACCRVSWLLWVYLLVMFLL 82
L FLLL+S + LV CC + L++Y LV FL+
Sbjct: 1 LNYFLLLLSHSRLVCLCCT*TTTLFIYSLVSFLV 102
>TC210903
Length = 687
Score = 25.0 bits (53), Expect(2) = 0.88
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 9/50 (18%)
Frame = +3
Query: 94 FAVTNKGAGEALSGRGYREYRLGDYSNWLQK---------RVNSTGNWNR 134
FA+T+ G+ + G+R Y GD++ L+ R N T WNR
Sbjct: 384 FAMTDSGSQLSSLVSGFRTYYGGDHAPTLRPNKTRLAAILRENDTIRWNR 533
Score = 23.9 bits (50), Expect(2) = 0.88
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = +1
Query: 68 VSWLLWVYLLVMFLLIVL 85
+SWLLW+ L V L+ +L
Sbjct: 337 LSWLLWISLHVQVLMYLL 390
>BG839888
Length = 707
Score = 29.3 bits (64), Expect = 1.9
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Frame = +2
Query: 53 LLLVSLAGLVGACCRVSWLLWVYLLV-MFLLIVLVFAFTIF 92
L+ SLA G C SWL+ + LL+ +F L+ L+ F F
Sbjct: 104 LIXYSLAVFSGPLCFXSWLMMLVLLLHLFXLLTLMCIFXFF 226
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.141 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,322,899
Number of Sequences: 63676
Number of extensions: 308982
Number of successful extensions: 2450
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 2406
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2436
length of query: 266
length of database: 12,639,632
effective HSP length: 96
effective length of query: 170
effective length of database: 6,526,736
effective search space: 1109545120
effective search space used: 1109545120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0041.4


