

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
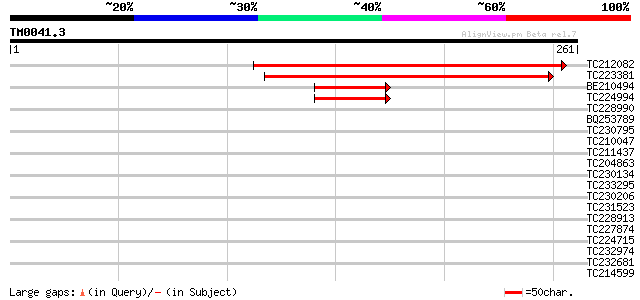
Query= TM0041.3
(261 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC212082 243 6e-65
TC223381 206 9e-54
BE210494 58 5e-09
TC224994 similar to GB|AAD39655.1|5103825|F9L1 ESTs gb|AA650895,... 58 5e-09
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 40 0.001
BQ253789 similar to GP|20805123|dbj P0039G05.13 {Oryza sativa (j... 40 0.001
TC230795 weakly similar to UP|O23299 (O23299) Carnitine racemase... 40 0.001
TC210047 weakly similar to UP|ROA1_HUMAN (P09651) Heterogeneous ... 38 0.004
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 38 0.004
TC204863 weakly similar to GB|AAM61695.1|21537354|AY085142 hydro... 37 0.007
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 37 0.009
TC233295 similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial (6%) 37 0.009
TC230206 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 37 0.009
TC231523 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 37 0.012
TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partia... 36 0.015
TC227874 weakly similar to UP|O22670 (O22670) Ag13 protein precu... 36 0.015
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 36 0.020
TC232974 weakly similar to GB|AAK54457.1|20384750|AY032998 DNA r... 36 0.020
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 35 0.026
TC214599 GB|AAA34124.1|170354|TOBUBI4A pentameric polyubiquitin ... 35 0.026
>TC212082
Length = 769
Score = 243 bits (620), Expect = 6e-65
Identities = 122/144 (84%), Positives = 129/144 (88%)
Frame = +1
Query: 113 LMALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVL 172
LMALIA+D+PGVDR+KC+KMAIVHDIAEAIVGDITP DGV K EKN+ EQEALDHMCKVL
Sbjct: 16 LMALIASDVPGVDRNKCIKMAIVHDIAEAIVGDITPLDGVSKAEKNQREQEALDHMCKVL 195
Query: 173 GGGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGK 232
GGGS A E+AELW EYEANSSPEAKFVKDLDKVEMILQALEYE EQGKDLDEFF STAGK
Sbjct: 196 GGGSTAMEIAELWMEYEANSSPEAKFVKDLDKVEMILQALEYEVEQGKDLDEFFWSTAGK 375
Query: 233 FQTEIGKAWASEIVSRRKKTNGSS 256
FQTE G AWASEIVSRRK +S
Sbjct: 376 FQTETGXAWASEIVSRRKNNK*TS 447
>TC223381
Length = 598
Score = 206 bits (524), Expect = 9e-54
Identities = 98/133 (73%), Positives = 117/133 (87%)
Frame = +2
Query: 118 AADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSR 177
A D+PG++R++C+K+A+VHDIAEAIVGDITP+DGVPK EK+R+E EAL+ MC++LGGG R
Sbjct: 2 AGDVPGLNRERCIKIALVHDIAEAIVGDITPSDGVPKAEKSRMELEALNKMCELLGGGMR 181
Query: 178 AKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEI 237
A+E+ ELW EYE NSS EA VKD DKVEMILQALEYE E GK LDEFF STAGKFQTEI
Sbjct: 182 AEEIKELWEEYENNSSVEANLVKDFDKVEMILQALEYEIEHGKVLDEFFLSTAGKFQTEI 361
Query: 238 GKAWASEIVSRRK 250
GK+WA+EI+SRRK
Sbjct: 362 GKSWAAEIISRRK 400
>BE210494
Length = 403
Score = 57.8 bits (138), Expect = 5e-09
Identities = 27/35 (77%), Positives = 28/35 (79%)
Frame = +2
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGG 175
AIVGDITP DG K EKN+ EQ LDHMCKVLGGG
Sbjct: 203 AIVGDITPLDGGSKAEKNQREQATLDHMCKVLGGG 307
>TC224994 similar to GB|AAD39655.1|5103825|F9L1 ESTs gb|AA650895, gb|AA720043
and gb|R29777 come from this gene. {Arabidopsis
thaliana;} , partial (72%)
Length = 1278
Score = 57.8 bits (138), Expect = 5e-09
Identities = 27/35 (77%), Positives = 28/35 (79%)
Frame = +1
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGG 175
AIVGDITP DG K EKN+ EQ LDHMCKVLGGG
Sbjct: 700 AIVGDITPLDGGSKAEKNQREQATLDHMCKVLGGG 804
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 40.0 bits (92), Expect = 0.001
Identities = 25/68 (36%), Positives = 38/68 (55%), Gaps = 8/68 (11%)
Frame = +2
Query: 16 SIPSSLSSTQLRFKSVT--------LSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAAS 67
S+PSS++S LRF + T LSP P T+A +SSP S +T+P S +S
Sbjct: 476 SLPSSIASPTLRFSTSTTTT*PENFLSPSLPCPSSATFTSAATSSPARSLPSTAPGSTSS 655
Query: 68 ATPSASSA 75
+PS +++
Sbjct: 656 TSPSPATS 679
Score = 27.3 bits (59), Expect = 7.2
Identities = 24/77 (31%), Positives = 36/77 (46%), Gaps = 4/77 (5%)
Frame = +2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSA---- 65
S+ F+S+ SS + + + S SPS A S +P S TSPPSA
Sbjct: 578 SATFTSAATSSPARSLPSTAPGSTSSTSPS------PATSSPAPSRPSSETSPPSASSTS 739
Query: 66 ASATPSASSAIDFLSIC 82
A+ TP+ +++ L C
Sbjct: 740 ATTTPTPAASRPRLETC 790
>BQ253789 similar to GP|20805123|dbj P0039G05.13 {Oryza sativa (japonica
cultivar-group)}, partial (4%)
Length = 293
Score = 40.0 bits (92), Expect = 0.001
Identities = 19/42 (45%), Positives = 26/42 (61%)
Frame = +2
Query: 49 GSSSPPSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKR 90
G P ++ + SA + S+SS IDFL++CHRLK TKR
Sbjct: 164 GFGEPINNKHASRDASAGLSASSSSSVIDFLTLCHRLKITKR 289
>TC230795 weakly similar to UP|O23299 (O23299) Carnitine racemase like
protein (AT4g14430/dl3255c), partial (47%)
Length = 749
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/64 (39%), Positives = 34/64 (53%), Gaps = 3/64 (4%)
Frame = +3
Query: 15 SSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASA---TPS 71
S +PSSL S + K P SP R + A +S+ P T+ PP++ASA TPS
Sbjct: 66 SYLPSSLRSPKSSPKPPPAPPSSPPRTADSSATASTSAGPRPPATSPPPASASAACPTPS 245
Query: 72 ASSA 75
A S+
Sbjct: 246 APSS 257
>TC210047 weakly similar to UP|ROA1_HUMAN (P09651) Heterogeneous nuclear
ribonucleoprotein A1 (Helix-destabilizing protein)
(Single-strand binding protein) (hnRNP core protein A1),
partial (12%)
Length = 520
Score = 38.1 bits (87), Expect = 0.004
Identities = 25/73 (34%), Positives = 40/73 (54%)
Frame = +3
Query: 2 ATATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTS 61
+T++R + +SS+P +LS T P SPS V + A+ + PP++S +S
Sbjct: 219 STSSRTRWRATPTSSLPCALSQT----------PTSPS-VSSSSAASAGTPPPTASAPSS 365
Query: 62 PPSAASATPSASS 74
PP+A S PS+ S
Sbjct: 366 PPTATSRRPSSYS 404
Score = 34.3 bits (77), Expect = 0.059
Identities = 24/56 (42%), Positives = 32/56 (56%)
Frame = +3
Query: 18 PSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSAS 73
P S SST R S T +P SPS T A+ SS+ P +S +TS + ATP++S
Sbjct: 102 PRSASST--RTASSTTTPPSPSSSPPPTPASSSSASPLTSSSTSSRTRWRATPTSS 263
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 38.1 bits (87), Expect = 0.004
Identities = 30/87 (34%), Positives = 44/87 (50%), Gaps = 13/87 (14%)
Frame = -1
Query: 9 LSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTT--AAGSSSPPSS---------- 56
+SS SSS SS S + S SP S S + + A+ S SPPSS
Sbjct: 608 VSSSSSSSSSSSSSPSSSSSSSPLSSPSSSSSPSSLPSPLASSSGSPPSSPSSPENSSSS 429
Query: 57 -SVTTSPPSAASATPSASSAIDFLSIC 82
S ++S PS++S++PS+SS++ C
Sbjct: 428 PSSSSSGPSSSSSSPSSSSSVSLFLFC 348
>TC204863 weakly similar to GB|AAM61695.1|21537354|AY085142
hydroxyproline-rich glycoprotein-like protein
{Arabidopsis thaliana;} , partial (30%)
Length = 786
Score = 37.4 bits (85), Expect = 0.007
Identities = 29/57 (50%), Positives = 34/57 (58%), Gaps = 1/57 (1%)
Frame = +2
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSS-VTTSPPSAASATPSASSA 75
S SST + S LS SPS T AA SPPSS+ +TT PP+A S TP ASS+
Sbjct: 278 SASSTTAKPLSTPLS-RSPSA--RSTRAAPLLSPPSSTTLTTPPPTARSMTPPASSS 439
Score = 29.3 bits (64), Expect = 1.9
Identities = 19/42 (45%), Positives = 27/42 (64%)
Frame = +2
Query: 33 LSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASS 74
LSP S + + A S +PP+SS ++S PSA++ T SASS
Sbjct: 362 LSPPSSTTLTTPPPTARSMTPPASS-SSSSPSASAWTRSASS 484
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 37.0 bits (84), Expect = 0.009
Identities = 42/143 (29%), Positives = 63/143 (43%)
Frame = -3
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SSLS L S + S SP ++++ SSS PSS TS S++S+
Sbjct: 456 SSSSSSSSSSSLSPPPLASGSSSESASSPPSSPEKSSSSSSSSSPSSPSFTSSSSSSSSV 277
Query: 70 PSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKC 129
AS + +C L GW + ++ P +I D L+A + D P R +
Sbjct: 276 SLAS--LFSSEVCFPLH-----GWSKGNL--PSTIGD----TKLLASVRRD-PVTTRKQA 139
Query: 130 VKMAIVHDIAEAIVGDITPTDGV 152
+A+ ++ PT V
Sbjct: 138 RDLAVARRVSSTKASTKEPTHKV 70
>TC233295 similar to UP|Q7KVQ0 (Q7KVQ0) CG4038-PA, partial (6%)
Length = 557
Score = 37.0 bits (84), Expect = 0.009
Identities = 27/66 (40%), Positives = 35/66 (52%), Gaps = 7/66 (10%)
Frame = +1
Query: 18 PSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASA-------TP 70
P S SST T SP S + ++ A+ +SS P SS TT PPS+ASA TP
Sbjct: 178 PPSRSSTS------TTSPPRSSTIPEVSLASPNSSTPCSSSTTRPPSSASAFAAPTLTTP 339
Query: 71 SASSAI 76
A+S +
Sbjct: 340 RATSPL 357
>TC230206 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (28%)
Length = 1305
Score = 37.0 bits (84), Expect = 0.009
Identities = 27/69 (39%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Frame = +1
Query: 8 LLSSLFSSSIPSSLSSTQLRFK---SVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPS 64
LL ++ PSS ++T R + T SP SP+R + A SSSPP S SPPS
Sbjct: 145 LLHGTTKTTKPSSCAATAARSSLAPTTTSSPTSPARPKFSPSTAPSSSPPCS--PNSPPS 318
Query: 65 AASATPSAS 73
A T + S
Sbjct: 319 ATPQTTTTS 345
>TC231523 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (32%)
Length = 878
Score = 36.6 bits (83), Expect = 0.012
Identities = 23/64 (35%), Positives = 36/64 (55%)
Frame = +2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS ++ PSS ++T R + SP T+AA S PS++ ++SPPS+ S+
Sbjct: 110 SSRTKTTRPSSCAATAARSSPAPTTTSSP------TSAATPRSSPSTAPSSSPPSSPSSP 271
Query: 70 PSAS 73
PSA+
Sbjct: 272 PSAT 283
>TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (19%)
Length = 692
Score = 36.2 bits (82), Expect = 0.015
Identities = 22/59 (37%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Frame = +2
Query: 17 IPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSS-SVTTSPPSAASATPSASS 74
+PS+ +S+ + SP S S V + + + SPPS+ S TT+PP A +TPS+S+
Sbjct: 338 VPSASTSST----TSAASPASASSVANTASVTTAPSPPSTPSTTTTPPPTARSTPSSSN 502
Score = 31.2 bits (69), Expect = 0.50
Identities = 32/94 (34%), Positives = 39/94 (41%), Gaps = 16/94 (17%)
Frame = +2
Query: 3 TATRVLLSSLF------SSSIPS---SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSS- 52
TATR +S SSS PS S S+T P SPS + T +G SS
Sbjct: 35 TATRRTTTSTLPPASPRSSSPPSSPPSPSTTPTSSAPANAPPSSPSASTRLQTPSGPSSD 214
Query: 53 ------PPSSSVTTSPPSAASATPSASSAIDFLS 80
P S+S +P + S PSAS A S
Sbjct: 215 ASTSPKPTSTSSRAAPSKSPSTWPSASLATSMSS 316
>TC227874 weakly similar to UP|O22670 (O22670) Ag13 protein precursor,
partial (21%)
Length = 894
Score = 36.2 bits (82), Expect = 0.015
Identities = 23/66 (34%), Positives = 36/66 (53%)
Frame = -2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SSLFS+ SSL+S F S+TL+ + S V +A S P ++S ++ S + +
Sbjct: 488 SSLFSAGASSSLASASASFSSLTLTSSAASGVLISAGSAFSFVPSAASCFSTVGSCVTVS 309
Query: 70 PSASSA 75
+ SSA
Sbjct: 308 DTTSSA 291
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 35.8 bits (81), Expect = 0.020
Identities = 23/57 (40%), Positives = 32/57 (55%), Gaps = 1/57 (1%)
Frame = +2
Query: 15 SSIPSSLSSTQLRFKSVTLSPHSPSRVFHM-TTAAGSSSPPSSSVTTSPPSAASATP 70
SS PS + T S SP +PS ++AA S+S PSSS +SPPS+ ++P
Sbjct: 707 SSSPSPSAPTSPAATSTPPSPSAPSSAATSPSSAASSTSSPSSSAPSSPPSSWPSSP 877
Score = 34.3 bits (77), Expect = 0.059
Identities = 22/46 (47%), Positives = 27/46 (57%), Gaps = 4/46 (8%)
Frame = +2
Query: 34 SPHSPSRVFHMTTAAGSSSPPS----SSVTTSPPSAASATPSASSA 75
SP S T+ A +S+PPS SS TSP SAAS+T S SS+
Sbjct: 701 SPSSSPSPSAPTSPAATSTPPSPSAPSSAATSPSSAASSTSSPSSS 838
Score = 27.3 bits (59), Expect = 7.2
Identities = 27/90 (30%), Positives = 40/90 (44%), Gaps = 17/90 (18%)
Frame = +2
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHS-----------PSRVFHMTTAAGSS 51
T TR L SS SS S Q T SP + PS + +++ S
Sbjct: 548 TPTR*RRLWLSSSQPSSSCSPAQAPASPTTSSPTTALPPLPASSPPPSPMHSPSSSPSPS 727
Query: 52 SPPSSSVTTSPPS------AASATPSASSA 75
+P S + T++PPS AA++ SA+S+
Sbjct: 728 APTSPAATSTPPSPSAPSSAATSPSSAASS 817
>TC232974 weakly similar to GB|AAK54457.1|20384750|AY032998 DNA repair
protein XRCC3 {Arabidopsis thaliana;} , partial (40%)
Length = 909
Score = 35.8 bits (81), Expect = 0.020
Identities = 28/70 (40%), Positives = 35/70 (50%), Gaps = 1/70 (1%)
Frame = +3
Query: 7 VLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP-PSA 65
+ S S + PS SS + + + +SPS A S PPSSS +SP PSA
Sbjct: 66 IAASPAASPAPPSPSSSARAVAARRSSASNSPSPPSSPPLTAVSPPPPSSSTPSSPSPSA 245
Query: 66 ASATPSASSA 75
ASAT A SA
Sbjct: 246 ASATSHAPSA 275
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 35.4 bits (80), Expect = 0.026
Identities = 27/75 (36%), Positives = 44/75 (58%)
Frame = -2
Query: 10 SSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASAT 69
SS SSS SS SS+ S + SP + + ++++ SSSPPS + ++S S++S++
Sbjct: 401 SSPCSSSSSSSSSSSSSSLSSSSGSPPRWTSLSSSSSSSSSSSPPSGASSSS--SSSSSS 228
Query: 70 PSASSAIDFLSICHR 84
S+SS+ D HR
Sbjct: 227 LSSSSSKDSRRRLHR 183
Score = 32.7 bits (73), Expect = 0.17
Identities = 27/73 (36%), Positives = 37/73 (49%)
Frame = -2
Query: 3 TATRVLLSSLFSSSIPSSLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSP 62
T R+ S S+S P S SS+ S + S T+ + SSS SSS SP
Sbjct: 440 TRGRIRYSVPKSASSPCSSSSSSSSSSSSSSLSSSSGSPPRWTSLSSSSSSSSSS---SP 270
Query: 63 PSAASATPSASSA 75
PS AS++ S+SS+
Sbjct: 269 PSGASSSSSSSSS 231
Score = 27.3 bits (59), Expect = 7.2
Identities = 12/37 (32%), Positives = 23/37 (61%)
Frame = -1
Query: 46 TAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLSIC 82
+++ SSS PSS+ ++S ++S++ S + F S C
Sbjct: 201 SSSSSSSSPSSTTSSSDSYSSSSSSSTKTVTSFSSTC 91
>TC214599 GB|AAA34124.1|170354|TOBUBI4A pentameric polyubiquitin {Nicotiana
sylvestris;} , partial (33%)
Length = 724
Score = 35.4 bits (80), Expect = 0.026
Identities = 28/78 (35%), Positives = 39/78 (49%), Gaps = 4/78 (5%)
Frame = +3
Query: 8 LLSSLFSSSIPSSLSSTQLRFKSVTLS----PHSPSRVFHMTTAAGSSSPPSSSVTTSPP 63
LLS SSS PS + LR+K++T S P S +R + SSSP SSS T P
Sbjct: 114 LLSRCRSSSRPSPERPSPLRWKALTPSTTSRPRSRTRKEFPRISNVSSSPESSSRTAVPS 293
Query: 64 SAASATPSASSAIDFLSI 81
++ S + F+S+
Sbjct: 294 PTTTSRRSQPFTLSFVSV 347
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,040,507
Number of Sequences: 63676
Number of extensions: 155788
Number of successful extensions: 4182
Number of sequences better than 10.0: 600
Number of HSP's better than 10.0 without gapping: 2833
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3747
length of query: 261
length of database: 12,639,632
effective HSP length: 95
effective length of query: 166
effective length of database: 6,590,412
effective search space: 1094008392
effective search space used: 1094008392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0041.3


