

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0040.4
(457 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
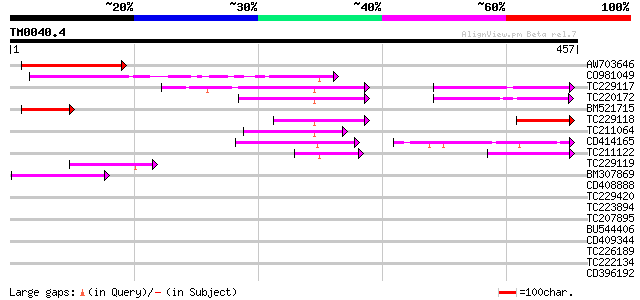
Score E
Sequences producing significant alignments: (bits) Value
AW703646 114 7e-26
CO981049 100 2e-21
TC229117 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization res... 74 2e-13
TC220172 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization res... 70 2e-12
BM521715 66 3e-11
TC229118 similar to UP|Q6R2U8 (Q6R2U8) Reduced vernalization res... 57 2e-08
TC211064 56 4e-08
CD414165 54 1e-07
TC211122 52 6e-07
TC229119 52 7e-07
BM307869 weakly similar to GP|20145851|emb auxin response factor... 43 3e-04
CD408888 similar to GP|20145851|emb auxin response factor 36 {Ar... 41 0.001
TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomai... 40 0.003
TC223894 37 0.024
TC207895 similar to UP|O81500 (O81500) F9D12.16 protein (At5g263... 32 0.60
BU544406 weakly similar to PIR|T39903|T399 serine-rich protein -... 30 1.7
CD409344 30 1.7
TC226189 similar to UP|Q7XL02 (Q7XL02) OJ000315_02.15 protein, p... 30 2.3
TC222134 similar to UP|RS14_CAEEL (P48150) 40S ribosomal protein... 30 2.3
CD396192 30 3.0
>AW703646
Length = 422
Score = 114 bits (286), Expect = 7e-26
Identities = 48/85 (56%), Positives = 62/85 (72%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
IPN F K+YG + N FLK P+G +WK+ K DG +WFQ GWK+FA+Y+SL HGH+++
Sbjct: 41 IPNKFVKKYGKRLQNTLFLKTPNGAEWKMILKKRDGKIWFQKGWKEFAEYHSLAHGHLLV 220
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPS 94
F + TSHF VHIFD S LEI YP+
Sbjct: 221 FRWDVTSHFQVHIFDLSALEIEYPT 295
>CO981049
Length = 665
Score = 99.8 bits (247), Expect = 2e-21
Identities = 74/255 (29%), Positives = 111/255 (43%), Gaps = 6/255 (2%)
Frame = -2
Query: 17 RYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVLFEYKDTS 76
+YG ++N FLK P+G +W++ K DG VWFQ GWK F +++SL HGH+++F+Y T
Sbjct: 661 KYGKHLSNTMFLKLPNGAEWRVNLEKRDGRVWFQEGWKKFVEHHSLAHGHLLVFKYDGTF 482
Query: 77 HFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPR 136
HF V IFD S EI YP ++ H +I + ++ P +C
Sbjct: 481 HFHVLIFDPSANEIDYPVNK---ANHKRVRISSEEIQ--------------PPTTCKTSG 353
Query: 137 KKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQSKF 196
K S +D H K+ D GR EG+ + EA
Sbjct: 352 NKRSNSNLQD----NAFHQ-----KVRDHKGRYESPSEGK----RNMEAAGSI------- 233
Query: 197 PSFMTVMKPAYVNGYSLHIPSFFAKKYLKNETDVLLQVLDGRTWSVSFNLGK------FN 250
SF MK + + +++P K Y+K + ++ R+W V K F+
Sbjct: 232 -SFTVRMKSS--SKQHMYLPKDSLKGYIKGGEQYVKLLVGERSWRVKLVHYKNRSSCFFS 62
Query: 251 AGWKKFTSVNNLKVG 265
A W F +LK G
Sbjct: 61 ANWPAFARXXDLKEG 17
Score = 38.1 bits (87), Expect = 0.008
Identities = 55/240 (22%), Positives = 90/240 (36%), Gaps = 14/240 (5%)
Frame = -2
Query: 209 NGYSLHIPS-FFAKKYLKNETDVLLQVLDGRTWSVSFNLGKFNAGWKKFTSVNNLKVGDV 267
N Y H+ + F K E V L+ DGR W F GWKKF ++L G +
Sbjct: 664 NKYGKHLSNTMFLKLPNGAEWRVNLEKRDGRVW--------FQEGWKKFVEHHSLAHGHL 509
Query: 268 CLFELNKSVPLSLKVLIFPLAEEPHSLPRSLVQGDRVNHINYGRFRYTESQNVLTSKCAE 327
+F+ + + VLIF + P VN N+ R R + + + C
Sbjct: 508 LVFKYDGT--FHFHVLIFDPSANEIDYP--------VNKANHKRVRISSEEIQPPTTCKT 359
Query: 328 T-IKRTSKLISPCPLKNAALEEANKFNSNN------------PFFIVNIAPDKNRDYRPR 374
+ KR++ + + ++ S + F + + K Y
Sbjct: 358 SGNKRSNSNLQDNAFHQKVRDHKGRYESPSEGKRNMEAAGSISFTVRMKSSSKQHMY--- 188
Query: 375 VPKLFIRKHFNDIQQSEQTVILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVG 434
+PK ++ + I+ EQ V L ++ VKL Y + S W FA+ L+ G
Sbjct: 187 LPKDSLKGY---IKGGEQYVKLLVGERSWRVKLVHYKNRSSCFFSANWPAFARXXDLKEG 17
>TC229117 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (38%)
Length = 904
Score = 73.6 bits (179), Expect = 2e-13
Identities = 54/177 (30%), Positives = 89/177 (49%), Gaps = 9/177 (5%)
Frame = +3
Query: 123 KTKLKSPMSCPKPRKKLRTSTSEDVGESPELHNLPK---QVKIEDDAGRTTECLEGEDLT 179
K + + + K KK R S E GE P N + + + + A + E+
Sbjct: 156 KLRYSNEETVNKTAKKKRKS--EPYGEEPSGENEEEAEMRYRFYESASARKRTVTAEERE 329
Query: 180 SKITEALNRARTFQSKFPSFMTVMKPAYV-NGYSLHIPSFFAKKYLKNETDVL-LQVLDG 237
A+N ++TF+ P V++P+Y+ G +++PS FA+K L + + LQ+ +G
Sbjct: 330 ----RAINASKTFEPTNPFCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGFIKLQLSNG 497
Query: 238 RTWSVSF----NLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEE 290
R WSV K + GW +FT NNL GDVC+FEL ++ + L+V +F + E+
Sbjct: 498 RQWSVRCLYRGGRAKLSQGWFEFTVENNLGEGDVCVFELLRTKEVVLQVTVFRVTED 668
Score = 51.2 bits (121), Expect = 1e-06
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 1/115 (0%)
Frame = +3
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ + F NPF V + P R +P F K+ N + + + +
Sbjct: 324 RERAINASKTFEPTNPFCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGFIKLQLSNGRQ 503
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
+ C + G ++LS+GW EF ++ L GD CVFEL+ ++ VL V ++R
Sbjct: 504 W----SVRCLYRGGRAKLSQGWFEFTVENNLGEGDVCVFELLRTKEVVLQVTVFR 656
>TC220172 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (39%)
Length = 798
Score = 70.5 bits (171), Expect = 2e-12
Identities = 40/112 (35%), Positives = 65/112 (57%), Gaps = 6/112 (5%)
Frame = +3
Query: 185 ALNRARTFQSKFPSFMTVMKPAYV-NGYSLHIPSFFAKKYLKNETDVL-LQVLDGRTWSV 242
A+N A+ F+ P V++P+Y+ G +++PS FA+K+L + + LQ+ +GR W V
Sbjct: 111 AINEAKAFEPSNPFCRVVLRPSYLYRGCIMYLPSCFAEKHLNGVSGFIKLQISNGRQWPV 290
Query: 243 SF----NLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEE 290
K + GW +F+ NNL GDVC+FEL + + L+V IF + E+
Sbjct: 291 RCLYKGGRAKLSQGWFEFSLENNLGEGDVCVFELLRMKEVVLQVTIFHVTED 446
Score = 60.1 bits (144), Expect = 2e-09
Identities = 39/114 (34%), Positives = 58/114 (50%), Gaps = 1/114 (0%)
Frame = +3
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ EA F +NPF V + P R +P F KH N + + I
Sbjct: 102 RERAINEAKAFEPSNPFCRVVLRPSYLYRGCIMYLPSCFAEKHLNGVSGFIKLQI--SNG 275
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIY 454
+ PV+ C K G ++LS+GW EF+ ++ L GD CVFEL+ ++ VL V I+
Sbjct: 276 RQWPVR--CLYKGGRAKLSQGWFEFSLENNLGEGDVCVFELLRMKEVVLQVTIF 431
>BM521715
Length = 277
Score = 66.2 bits (160), Expect = 3e-11
Identities = 24/43 (55%), Positives = 34/43 (78%)
Frame = +2
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNG 52
+P FT++YG M+NP FLKP DGT+WKI+++K G++WFQ G
Sbjct: 149 LPKKFTRKYGDGMSNPVFLKPADGTEWKIHYTKHGGEIWFQKG 277
>TC229118 similar to UP|Q6R2U8 (Q6R2U8) Reduced vernalization response 1,
partial (17%)
Length = 470
Score = 56.6 bits (135), Expect = 2e-08
Identities = 32/83 (38%), Positives = 49/83 (58%), Gaps = 5/83 (6%)
Frame = +3
Query: 213 LHIPSFFAKKYLKNETDVL-LQVLDGRTWSVSF----NLGKFNAGWKKFTSVNNLKVGDV 267
+++PS FA+K L + + LQ+ +GR WSV K + GW +FT NNL GDV
Sbjct: 9 MYLPSTFAEKNLNGVSGFIKLQLSNGRQWSVRCLYRGGRAKLSQGWFEFTVENNLGEGDV 188
Query: 268 CLFELNKSVPLSLKVLIFPLAEE 290
C+FEL + + L+V +F + E+
Sbjct: 189 CVFELLRMKEVVLQVTVFRVTED 257
Score = 47.4 bits (111), Expect = 1e-05
Identities = 20/47 (42%), Positives = 31/47 (65%)
Frame = +3
Query: 409 CYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
C + G ++LS+GW EF ++ L GD CVFEL+ ++ VL V ++R
Sbjct: 105 CLYRGGRAKLSQGWFEFTVENNLGEGDVCVFELLRMKEVVLQVTVFR 245
>TC211064
Length = 706
Score = 55.8 bits (133), Expect = 4e-08
Identities = 34/93 (36%), Positives = 50/93 (53%), Gaps = 9/93 (9%)
Frame = +3
Query: 189 ARTFQSKFPSFMTVMKPAYVNG-YSLHIPSFFAKKYLKN-ETDVLLQVLDGRTWSVSF-- 244
A+ F S FPSF+ +MK V+G Y+L IP F+ +L +T+V L+ G W+V+
Sbjct: 231 AQAFTSPFPSFVKIMKKFNVSGSYTLKIPYQFSAAHLPTYKTEVTLRNSRGECWTVNSVP 410
Query: 245 -----NLGKFNAGWKKFTSVNNLKVGDVCLFEL 272
+ F GW F N++ GD C+FEL
Sbjct: 411 DAKGRTVHTFCGGWMAFVRDNDINFGDTCIFEL 509
Score = 32.0 bits (71), Expect = 0.60
Identities = 11/34 (32%), Positives = 21/34 (61%)
Frame = +3
Query: 421 GWVEFAKKSKLEVGDACVFELINKEDPVLDVHIY 454
GW+ F + + + GD C+FEL+ + ++H+Y
Sbjct: 447 GWMAFVRDNDINFGDTCIFELVAQ----CEMHVY 536
>CD414165
Length = 623
Score = 54.3 bits (129), Expect = 1e-07
Identities = 45/159 (28%), Positives = 75/159 (46%), Gaps = 13/159 (8%)
Frame = -1
Query: 310 GRFRYTESQNVLTSKCAETIKRTSKLIS----PCPLKNAALEE------ANKFNSNNPFF 359
G F + ES+ S C E + ++K + P PL + E A F S NP +
Sbjct: 599 GSFLHHESK----SSCQEDLPFSNKALLSKDFPKPLSSIKFETSEACKLAESFTSRNPHW 432
Query: 360 IVNIAPDKNRDYRP-RVPKLFIRKHFNDIQQSEQTVILQCEKKLCPVKLTC--YPKKGPS 416
+ D+ + F K+ ++ + Q ++ E K V++TC YP K +
Sbjct: 431 KHLMTKCNVEDHCTLHIATQFACKYIPEVVK--QIILWNSEGKFWEVEVTCLRYPNKRYT 258
Query: 417 RLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
R + GW +F + +KL GD C+FEL +++ + VHI+R
Sbjct: 257 RFTTGWGKFVRDNKLMPGDTCIFEL--EDENHMSVHIFR 147
Score = 50.4 bits (119), Expect = 2e-06
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 10/110 (9%)
Frame = -1
Query: 183 TEALNRARTFQSKFPSFMTVMKPAYVNGY-SLHIPSFFAKKYLKNET-DVLLQVLDGRTW 240
+EA A +F S+ P + +M V + +LHI + FA KY+ ++L +G+ W
Sbjct: 482 SEACKLAESFTSRNPHWKHLMTKCNVEDHCTLHIATQFACKYIPEVVKQIILWNSEGKFW 303
Query: 241 SVSFNL--------GKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKV 282
V +F GW KF N L GD C+FEL +S+ +
Sbjct: 302 EVEVTCLRYPNKRYTRFTTGWGKFVRDNKLMPGDTCIFELEDENHMSVHI 153
Score = 33.5 bits (75), Expect = 0.21
Identities = 23/83 (27%), Positives = 34/83 (40%), Gaps = 6/83 (7%)
Frame = -1
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKI------YHSKTDGDVWFQNGWKDFAKYYSLD 63
I F +Y ++ L +G W++ Y +K F GW F + L
Sbjct: 383 IATQFACKYIPEVVKQIILWNSEGKFWEVEVTCLRYPNKRY--TRFTTGWGKFVRDNKLM 210
Query: 64 HGHMVLFEYKDTSHFGVHIFDKS 86
G +FE +D +H VHIF S
Sbjct: 209 PGDTCIFELEDENHMSVHIFRTS 141
>TC211122
Length = 640
Score = 52.0 bits (123), Expect = 6e-07
Identities = 27/62 (43%), Positives = 32/62 (51%), Gaps = 6/62 (9%)
Frame = +1
Query: 230 VLLQVLDGRTWSVSFNLGK------FNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVL 283
+ LQVL GR W + + K F W F NNLKVGDVC+FEL L+ V
Sbjct: 25 ISLQVLSGRIWPAKYQIHKQKTAIRFKLSWNAFVKDNNLKVGDVCIFELVHGTKLTFLVH 204
Query: 284 IF 285
IF
Sbjct: 205 IF 210
Score = 44.3 bits (103), Expect = 1e-04
Identities = 22/71 (30%), Positives = 36/71 (49%), Gaps = 1/71 (1%)
Frame = +1
Query: 386 DIQQSEQTVILQC-EKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINK 444
D+ + + + LQ ++ P K + +K R W F K + L+VGD C+FEL++
Sbjct: 1 DLHKKRRLISLQVLSGRIWPAKYQIHKQKTAIRFKLSWNAFVKDNNLKVGDVCIFELVHG 180
Query: 445 EDPVLDVHIYR 455
VHI+R
Sbjct: 181 TKLTFLVHIFR 213
>TC229119
Length = 455
Score = 51.6 bits (122), Expect = 7e-07
Identities = 30/80 (37%), Positives = 48/80 (59%), Gaps = 9/80 (11%)
Frame = +2
Query: 49 FQNGWKDFAKYYSLDHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPS-HENQDE------- 100
F +GW+DF ++YS+ G+ ++F Y+ S F VHIF+ ST E+ Y S N++E
Sbjct: 5 FVDGWQDFVQHYSIGVGYFLVFMYEGNSSFIVHIFNLSTSEVNYQSAMRNRNEGSCLANY 184
Query: 101 EHNPDQIDD-DSVEILDKIP 119
H D ++D DS+++ D P
Sbjct: 185 HHIFD*MEDVDSLDLSDLSP 244
>BM307869 weakly similar to GP|20145851|emb auxin response factor 36
{Arabidopsis thaliana}, partial (8%)
Length = 437
Score = 42.7 bits (99), Expect = 3e-04
Identities = 21/79 (26%), Positives = 36/79 (44%)
Frame = +2
Query: 2 LMESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYS 61
+M + + +P F+ + LK P G W I + D ++F +GW+ F K +
Sbjct: 200 IMINTLLALPKAFSDNLKKKLPENVTLKGPGGVVWNIGMTTRDDTLYFAHGWEQFVKDHC 379
Query: 62 LDHGHMVLFEYKDTSHFGV 80
L ++F+Y S F V
Sbjct: 380 LKENDFLVFKYNGESQFDV 436
>CD408888 similar to GP|20145851|emb auxin response factor 36 {Arabidopsis
thaliana}, partial (6%)
Length = 597
Score = 41.2 bits (95), Expect = 0.001
Identities = 32/97 (32%), Positives = 48/97 (48%), Gaps = 10/97 (10%)
Frame = -1
Query: 197 PSFMTVMKPAYVNG-YSLHIPSFFAKKYLKNETDVLLQVLDGRTWS-VSFNLGKFNA--- 251
P + +V+ P + Y+L K NE +L + G++W V + K A
Sbjct: 567 PYWHSVLSPTNLQPRYNLMPGMNLRSKLPSNEVPTIL-IYGGKSWDMVYYGQSKQKAFGV 391
Query: 252 -GWKKFTSVNNLKVGDVCLFEL----NKSVPLSLKVL 283
GWKKF N L+VGD C+FEL +K V L +++L
Sbjct: 390 TGWKKFVIDNCLRVGDACIFELMESSDKKVILEVQIL 280
Score = 38.1 bits (87), Expect = 0.008
Identities = 18/38 (47%), Positives = 24/38 (62%), Gaps = 2/38 (5%)
Frame = -1
Query: 421 GWVEFAKKSKLEVGDACVFELINKEDP--VLDVHIYRG 456
GW +F + L VGDAC+FEL+ D +L+V I RG
Sbjct: 387 GWKKFVIDNCLRVGDACIFELMESSDKKVILEVQILRG 274
>TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomain-containing
transcription factor Nkx6.1 {Mus musculus;} , partial
(8%)
Length = 870
Score = 39.7 bits (91), Expect = 0.003
Identities = 31/103 (30%), Positives = 48/103 (46%), Gaps = 8/103 (7%)
Frame = +2
Query: 177 DLTSKITEALNRARTFQSKFPSFMTVMKP--AYVNGYSLHIPSFFAKKYLKNETDVLLQV 234
D +K+ E+L +A + S VMKP AY + ++ K + DV+L++
Sbjct: 188 DDETKMIESLAKAACTED---SIYVVMKPSHAYKRFFVSMRGTWIGKHISPSSQDVILRM 358
Query: 235 LDGRTWSVSF------NLGKFNAGWKKFTSVNNLKVGDVCLFE 271
G W + N G GWK F+ NNL+ GD C+F+
Sbjct: 359 GKGE-WIARYSYNNIRNNGGLTGGWKHFSLDNNLEEGDACVFK 484
Score = 35.0 bits (79), Expect = 0.070
Identities = 26/79 (32%), Positives = 40/79 (49%), Gaps = 2/79 (2%)
Frame = +2
Query: 379 FIRKHFNDIQQSEQTVILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACV 438
+I KH I S Q VIL+ K + + + L+ GW F+ + LE GDACV
Sbjct: 308 WIGKH---ISPSSQDVILRMGKGEWIARYSYNNIRNNGGLTGGWKHFSLDNNLEEGDACV 478
Query: 439 FELINKEDP--VLDVHIYR 455
F+ + + V+D+ I+R
Sbjct: 479 FKPAGQINNTFVIDMSIFR 535
>TC223894
Length = 702
Score = 36.6 bits (83), Expect = 0.024
Identities = 17/39 (43%), Positives = 24/39 (60%), Gaps = 3/39 (7%)
Frame = +2
Query: 420 KGWVEFAKKSKLEVGDACVFELINKEDPV---LDVHIYR 455
KGW F +++K+E D C E+I+ ED V L VH+ R
Sbjct: 443 KGWASFCRRNKIERNDTCFCEVISGEDQVVKTLRVHVAR 559
>TC207895 similar to UP|O81500 (O81500) F9D12.16 protein (At5g26330), partial
(53%)
Length = 858
Score = 32.0 bits (71), Expect = 0.60
Identities = 33/143 (23%), Positives = 52/143 (36%), Gaps = 14/143 (9%)
Frame = +3
Query: 233 QVLDGRTWSVSFNLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKV--LIFPLAEE 290
+V D W++ N+ +KK+ + N +VGD +FE N ++V ++
Sbjct: 207 KVGDSAGWTIIGNID-----YKKWAATKNFQVGDTIIFEYNAKFHNVMRVTHAMYKSCNA 371
Query: 291 PHSLPRSLVQGDRVNHINYGRFRY------------TESQNVLTSKCAETIKRTSKLISP 338
L D + NYG + NV+ A TS + SP
Sbjct: 372 SSPLTTMSTGNDTIKITNYGHHFFLCGIPGHCQAGQKVDINVVKVSAAAAPSPTSAMASP 551
Query: 339 CPLKNAALEEANKFNSNNPFFIV 361
P N A N+ PF +V
Sbjct: 552 VPPANV---PAPSPNNAAPFIVV 611
>BU544406 weakly similar to PIR|T39903|T399 serine-rich protein - fission
yeast (Schizosaccharomyces pombe), partial (5%)
Length = 696
Score = 30.4 bits (67), Expect = 1.7
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 3/39 (7%)
Frame = -3
Query: 420 KGWVEFAKKSKLEVGDACVFELINKEDP---VLDVHIYR 455
KGW F +++++E D C E+I+ E+ L VH+ R
Sbjct: 304 KGWDSFCRRNEIERDDTCFCEVISGEEQGVRTLRVHVAR 188
>CD409344
Length = 438
Score = 30.4 bits (67), Expect = 1.7
Identities = 20/80 (25%), Positives = 34/80 (42%), Gaps = 2/80 (2%)
Frame = -2
Query: 233 QVLDGRTWSVSFNLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVL--IFPLAEE 290
+V D W++ N+ +KK+ + N +VGD +FE N ++V ++
Sbjct: 356 KVGDSAGWTIIGNID-----YKKWAATKNFQVGDTIIFEYNAKFHNVMRVTHGMYKSCNA 192
Query: 291 PHSLPRSLVQGDRVNHINYG 310
L R D + NYG
Sbjct: 191 SSPLTRMSTGNDTIKITNYG 132
>TC226189 similar to UP|Q7XL02 (Q7XL02) OJ000315_02.15 protein, partial (81%)
Length = 1145
Score = 30.0 bits (66), Expect = 2.3
Identities = 15/40 (37%), Positives = 21/40 (52%)
Frame = -2
Query: 117 KIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVGESPELHNL 156
K+PPC L P+S + RK R+ S +PEL +L
Sbjct: 520 KLPPCWTGHLPCPLSFSRHRKSQRSLPSSQQSFAPELKDL 401
>TC222134 similar to UP|RS14_CAEEL (P48150) 40S ribosomal protein S14,
partial (80%)
Length = 471
Score = 30.0 bits (66), Expect = 2.3
Identities = 19/61 (31%), Positives = 30/61 (49%)
Frame = +2
Query: 122 KKTKLKSPMSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSK 181
+K+KLK P+S PR+ R ++ V S + +PK VK + T G L+S
Sbjct: 17 QKSKLKLPLSKESPRRVRRRVMTKKVPASKSSNWVPKSVKPNSSSVSLTSSPPGTILSST 196
Query: 182 I 182
+
Sbjct: 197 L 199
>CD396192
Length = 694
Score = 29.6 bits (65), Expect = 3.0
Identities = 11/21 (52%), Positives = 17/21 (80%)
Frame = +1
Query: 58 KYYSLDHGHMVLFEYKDTSHF 78
+Y+SL HGH++LF+ K T +F
Sbjct: 91 EYHSLSHGHLLLFKCKVTCNF 153
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,252,064
Number of Sequences: 63676
Number of extensions: 334172
Number of successful extensions: 1696
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1651
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1692
length of query: 457
length of database: 12,639,632
effective HSP length: 100
effective length of query: 357
effective length of database: 6,272,032
effective search space: 2239115424
effective search space used: 2239115424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0040.4


