

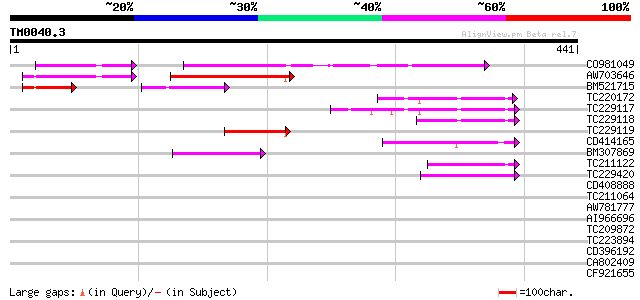
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0040.3
(441 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CO981049 113 1e-25
AW703646 112 3e-25
BM521715 69 5e-12
TC220172 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization res... 67 2e-11
TC229117 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization res... 60 3e-09
TC229118 similar to UP|Q6R2U8 (Q6R2U8) Reduced vernalization res... 53 3e-07
TC229119 49 3e-06
CD414165 48 1e-05
BM307869 weakly similar to GP|20145851|emb auxin response factor... 46 4e-05
TC211122 42 4e-04
TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomai... 42 6e-04
CD408888 similar to GP|20145851|emb auxin response factor 36 {Ar... 37 0.014
TC211064 36 0.030
AW781777 35 0.088
AI966696 31 1.3
TC209872 30 2.2
TC223894 30 2.2
CD396192 30 2.2
CA802409 similar to GP|24198299|gb| Strictosidine synthase precu... 30 2.2
CF921655 30 2.2
>CO981049
Length = 665
Score = 113 bits (283), Expect = 1e-25
Identities = 73/238 (30%), Positives = 112/238 (46%)
Frame = -2
Query: 136 KYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGHLVLFEFKDAS 195
KY +SN +FLK P+ W ++ K D WFQ+GWK+FV+++SL HGHL++F++
Sbjct: 661 KYGKHLSNTMFLKLPNGAEWRVNLEKRDGRVWFQEGWKKFVEHHSLAHGHLLVFKYDGTF 482
Query: 196 HFGVHIFDNSTLEIEYPSNENQGEEHNPDQIGDEILLPKKTKPKSPMSLSRPCKKLRTGK 255
HF V IFD S EI+YP N+ H +I E + P T CK +G
Sbjct: 481 HFHVLIFDPSANEIDYPVNK---ANHKRVRISSEEIQPPTT-----------CK--TSGN 350
Query: 256 SKDVGKSPELRKRKKVQVEGGEPSSLGPHFTAQGEARKYTPKNPSFTVSLKPVIKLAYRP 315
+ + +KV+ G S ++G+ + SFTV +K K
Sbjct: 349 KRSNSNLQDNAFHQKVRDHKGRYES-----PSEGKRNMEAAGSISFTVRMKSSSK--QHM 191
Query: 316 PVPISFIREYLNEKKQTIKLKIEENLWPVEFIYYPEHGSGKLSKGWIQFAKESKLVGG 373
+P ++ Y+ +Q +KL + E W V+ ++Y S S W FA+ L G
Sbjct: 190 YLPKDSLKGYIKGGEQYVKLLVGERSWRVKLVHYKNRSSCFFSANWPAFARXXDLKEG 17
Score = 62.8 bits (151), Expect = 3e-10
Identities = 30/78 (38%), Positives = 46/78 (58%)
Frame = -2
Query: 21 GDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMVWHLVLFEFKG 80
G ++SN +FLK + W +N K+D +W Q W++FV+ + L G HL++F++ G
Sbjct: 655 GKHLSNTMFLKLPNGAEWRVNLEKRDGRVWFQEGWKKFVEHHSLAHG----HLLVFKYDG 488
Query: 81 PSKFEVCIFDKNALEIKY 98
F V IFD +A EI Y
Sbjct: 487 TFHFHVLIFDPSANEIDY 434
>AW703646
Length = 422
Score = 112 bits (280), Expect = 3e-25
Identities = 52/109 (47%), Positives = 69/109 (62%), Gaps = 13/109 (11%)
Frame = +2
Query: 126 ELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGH 185
+L++PN F+KKY + N +FLK P+ W++ K D WFQKGWKEF +Y+SL HGH
Sbjct: 32 KLMIPNKFVKKYGKRLQNTLFLKTPNGAEWKMILKKRDGKIWFQKGWKEFAEYHSLAHGH 211
Query: 186 LVLFEFKDASHFGVHIFDNSTLEIEYPS-------------NENQGEEH 221
L++F + SHF VHIFD S LEIEYP+ NE+ G+EH
Sbjct: 212 LLVFRWDVTSHFQVHIFDLSALEIEYPTEIIKGKTASNRKGNESPGDEH 358
Score = 68.6 bits (166), Expect = 5e-12
Identities = 35/88 (39%), Positives = 52/88 (58%)
Frame = +2
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
+PN F +KY G + N +FLK + W++ K+D IW Q W+EF + + L G
Sbjct: 41 IPNKFVKKY-GKRLQNTLFLKTPNGAEWKMILKKRDGKIWFQKGWKEFAEYHSLAHG--- 208
Query: 71 WHLVLFEFKGPSKFEVCIFDKNALEIKY 98
HL++F + S F+V IFD +ALEI+Y
Sbjct: 209 -HLLVFRWDVTSHFQVHIFDLSALEIEY 289
>BM521715
Length = 277
Score = 68.6 bits (166), Expect = 5e-12
Identities = 34/69 (49%), Positives = 41/69 (59%)
Frame = +2
Query: 103 NKQIPNSATRFFRIITPPCLEAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKH 162
NK A F +II L A ++LP F +KY GMSNPVFLK D T W+IH++KH
Sbjct: 74 NKHPLPPAVHFVKIILTTSL-ADGILLPKKFTRKYGDGMSNPVFLKPADGTEWKIHYTKH 250
Query: 163 DSYFWFQKG 171
WFQKG
Sbjct: 251 GGEIWFQKG 277
Score = 57.0 bits (136), Expect = 2e-08
Identities = 25/42 (59%), Positives = 30/42 (70%)
Frame = +2
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQ 52
LP FTRKY GD MSNPVFLKP D W+I+++K +IW Q
Sbjct: 149 LPKKFTRKY-GDGMSNPVFLKPADGTEWKIHYTKHGGEIWFQ 271
>TC220172 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (39%)
Length = 798
Score = 66.6 bits (161), Expect = 2e-11
Identities = 46/113 (40%), Positives = 61/113 (53%), Gaps = 4/113 (3%)
Frame = +3
Query: 287 AQGEARKYTPKNPSFTVSLKPVIKLAYRPPV---PISFIREYLNEKKQTIKLKIEENL-W 342
A EA+ + P NP V L+P YR + P F ++LN IKL+I W
Sbjct: 111 AINEAKAFEPSNPFCRVVLRP--SYLYRGCIMYLPSCFAEKHLNGVSGFIKLQISNGRQW 284
Query: 343 PVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIF 395
PV +Y + G KLS+GW +F+ E+ L GDVCVFEL+ E VL+V IF
Sbjct: 285 PVRCLY--KGGRAKLSQGWFEFSLENNLGEGDVCVFELLRMKE--VVLQVTIF 431
Score = 31.2 bits (69), Expect = 0.97
Identities = 19/80 (23%), Positives = 34/80 (41%), Gaps = 2/80 (2%)
Frame = +3
Query: 113 FFRIITPPCL--EAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQK 170
F R++ P + LP+ F +K+ G+S + L+ + W + +
Sbjct: 150 FCRVVLRPSYLYRGCIMYLPSCFAEKHLNGVSGFIKLQISNGRQWPVRCLYKGGRAKLSQ 329
Query: 171 GWKEFVKYYSLDHGHLVLFE 190
GW EF +L G + +FE
Sbjct: 330 GWFEFSLENNLGEGDVCVFE 389
>TC229117 similar to UP|Q8L3W1 (Q8L3W1) Reduced vernalization response 1,
partial (38%)
Length = 904
Score = 59.7 bits (143), Expect = 3e-09
Identities = 56/175 (32%), Positives = 76/175 (43%), Gaps = 28/175 (16%)
Frame = +3
Query: 250 KLRTGKSKDVGKSPELRKRKKVQVEGGEPSS-------LGPHFTAQGEARKYT------- 295
KLR + V K+ +K++K + G EPS + F ARK T
Sbjct: 156 KLRYSNEETVNKTA--KKKRKSEPYGEEPSGENEEEAEMRYRFYESASARKRTVTAEERE 329
Query: 296 ----------PKNPSFTVSLKPVIKLAYRPPV---PISFIREYLNEKKQTIKLKIEENL- 341
P NP V L+P YR + P F + LN IKL++
Sbjct: 330 RAINASKTFEPTNPFCRVVLRP--SYLYRGCIMYLPSCFAEKNLNGVSGFIKLQLSNGRQ 503
Query: 342 WPVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIFK 396
W V +Y G KLS+GW +F E+ L GDVCVFEL+ E VL+V +F+
Sbjct: 504 WSVRCLY--RGGRAKLSQGWFEFTVENNLGEGDVCVFELLRTKE--VVLQVTVFR 656
Score = 31.2 bits (69), Expect = 0.97
Identities = 19/80 (23%), Positives = 33/80 (40%), Gaps = 2/80 (2%)
Frame = +3
Query: 113 FFRIITPPCL--EAAELILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQK 170
F R++ P + LP+ F +K G+S + L+ + W + +
Sbjct: 372 FCRVVLRPSYLYRGCIMYLPSCFAEKNLNGVSGFIKLQLSNGRQWSVRCLYRGGRAKLSQ 551
Query: 171 GWKEFVKYYSLDHGHLVLFE 190
GW EF +L G + +FE
Sbjct: 552 GWFEFTVENNLGEGDVCVFE 611
>TC229118 similar to UP|Q6R2U8 (Q6R2U8) Reduced vernalization response 1,
partial (17%)
Length = 470
Score = 52.8 bits (125), Expect = 3e-07
Identities = 32/81 (39%), Positives = 45/81 (55%), Gaps = 1/81 (1%)
Frame = +3
Query: 317 VPISFIREYLNEKKQTIKLKIEENL-WPVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDV 375
+P +F + LN IKL++ W V +Y G KLS+GW +F E+ L GDV
Sbjct: 15 LPSTFAEKNLNGVSGFIKLQLSNGRQWSVRCLY--RGGRAKLSQGWFEFTVENNLGEGDV 188
Query: 376 CVFELINKDEENPVLEVHIFK 396
CVFEL+ E VL+V +F+
Sbjct: 189 CVFELLRMKE--VVLQVTVFR 245
Score = 31.2 bits (69), Expect = 0.97
Identities = 16/64 (25%), Positives = 28/64 (43%)
Frame = +3
Query: 127 LILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGHL 186
+ LP+ F +K G+S + L+ + W + +GW EF +L G +
Sbjct: 9 MYLPSTFAEKNLNGVSGFIKLQLSNGRQWSVRCLYRGGRAKLSQGWFEFTVENNLGEGDV 188
Query: 187 VLFE 190
+FE
Sbjct: 189 CVFE 200
>TC229119
Length = 455
Score = 49.3 bits (116), Expect = 3e-06
Identities = 23/54 (42%), Positives = 36/54 (66%), Gaps = 3/54 (5%)
Frame = +2
Query: 168 FQKGWKEFVKYYSLDHGHLVLFEFKDASHFGVHIFDNSTLEIEYPS---NENQG 218
F GW++FV++YS+ G+ ++F ++ S F VHIF+ ST E+ Y S N N+G
Sbjct: 5 FVDGWQDFVQHYSIGVGYFLVFMYEGNSSFIVHIFNLSTSEVNYQSAMRNRNEG 166
>CD414165
Length = 623
Score = 47.8 bits (112), Expect = 1e-05
Identities = 33/110 (30%), Positives = 54/110 (49%), Gaps = 4/110 (3%)
Frame = -1
Query: 291 ARKYTPKNPSFT-VSLKPVIKLAYRPPVPISFIREYLNEK-KQTIKLKIEENLWPVEF-- 346
A +T +NP + + K ++ + F +Y+ E KQ I E W VE
Sbjct: 464 AESFTSRNPHWKHLMTKCNVEDHCTLHIATQFACKYIPEVVKQIILWNSEGKFWEVEVTC 285
Query: 347 IYYPEHGSGKLSKGWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIFK 396
+ YP + + GW +F +++KL+ GD C+FEL E+ + VHIF+
Sbjct: 284 LRYPNKRYTRFTTGWGKFVRDNKLMPGDTCIFEL----EDENHMSVHIFR 147
Score = 38.9 bits (89), Expect = 0.005
Identities = 23/77 (29%), Positives = 34/77 (43%), Gaps = 4/77 (5%)
Frame = -1
Query: 133 FMKKYSGGMSNPVFLKAPDNTSWEIHWS----KHDSYFWFQKGWKEFVKYYSLDHGHLVL 188
F KY + + L + WE+ + + Y F GW +FV+ L G +
Sbjct: 371 FACKYIPEVVKQIILWNSEGKFWEVEVTCLRYPNKRYTRFTTGWGKFVRDNKLMPGDTCI 192
Query: 189 FEFKDASHFGVHIFDNS 205
FE +D +H VHIF S
Sbjct: 191 FELEDENHMSVHIFRTS 141
>BM307869 weakly similar to GP|20145851|emb auxin response factor 36
{Arabidopsis thaliana}, partial (8%)
Length = 437
Score = 45.8 bits (107), Expect = 4e-05
Identities = 22/73 (30%), Positives = 32/73 (43%)
Frame = +2
Query: 127 LILPNGFMKKYSGGMSNPVFLKAPDNTSWEIHWSKHDSYFWFQKGWKEFVKYYSLDHGHL 186
L LP F + V LK P W I + D +F GW++FVK + L
Sbjct: 218 LALPKAFSDNLKKKLPENVTLKGPGGVVWNIGMTTRDDTLYFAHGWEQFVKDHCLKENDF 397
Query: 187 VLFEFKDASHFGV 199
++F++ S F V
Sbjct: 398 LVFKYNGESQFDV 436
Score = 37.0 bits (84), Expect = 0.018
Identities = 22/76 (28%), Positives = 38/76 (49%)
Frame = +2
Query: 11 LPNAFTRKYSGDNMSNPVFLKPLDNISWEINWSKQDSDIWLQNVWEEFVKLYYLDQGHMV 70
LP AF+ N V LK + W I + +D ++ + WE+FVK + L +
Sbjct: 224 LPKAFSDNLKKKLPEN-VTLKGPGGVVWNIGMTTRDDTLYFAHGWEQFVKDHCLKEN--- 391
Query: 71 WHLVLFEFKGPSKFEV 86
++F++ G S+F+V
Sbjct: 392 -DFLVFKYNGESQFDV 436
>TC211122
Length = 640
Score = 42.4 bits (98), Expect = 4e-04
Identities = 21/72 (29%), Positives = 41/72 (56%), Gaps = 1/72 (1%)
Frame = +1
Query: 326 LNEKKQTIKLKI-EENLWPVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDVCVFELINKD 384
L++K++ I L++ +WP ++ + + + + W F K++ L GDVC+FEL++
Sbjct: 4 LHKKRRLISLQVLSGRIWPAKYQIHKQKTAIRFKLSWNAFVKDNNLKVGDVCIFELVHGT 183
Query: 385 EENPVLEVHIFK 396
+ VHIF+
Sbjct: 184 K--LTFLVHIFR 213
>TC229420 homologue to GB|AAG30415.1|11118687|AF291666 homeodomain-containing
transcription factor Nkx6.1 {Mus musculus;} , partial
(8%)
Length = 870
Score = 42.0 bits (97), Expect = 6e-04
Identities = 19/77 (24%), Positives = 41/77 (52%)
Frame = +2
Query: 320 SFIREYLNEKKQTIKLKIEENLWPVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDVCVFE 379
++I ++++ Q + L++ + W + Y +G L+ GW F+ ++ L GD CVF+
Sbjct: 305 TWIGKHISPSSQDVILRMGKGEWIARYSYNNIRNNGGLTGGWKHFSLDNNLEEGDACVFK 484
Query: 380 LINKDEENPVLEVHIFK 396
+ V+++ IF+
Sbjct: 485 PAGQINNTFVIDMSIFR 535
>CD408888 similar to GP|20145851|emb auxin response factor 36 {Arabidopsis
thaliana}, partial (6%)
Length = 597
Score = 37.4 bits (85), Expect = 0.014
Identities = 15/38 (39%), Positives = 24/38 (62%)
Frame = -1
Query: 360 GWIQFAKESKLVGGDVCVFELINKDEENPVLEVHIFKG 397
GW +F ++ L GD C+FEL+ ++ +LEV I +G
Sbjct: 387 GWKKFVIDNCLRVGDACIFELMESSDKKVILEVQILRG 274
>TC211064
Length = 706
Score = 36.2 bits (82), Expect = 0.030
Identities = 42/172 (24%), Positives = 68/172 (39%), Gaps = 11/172 (6%)
Frame = +3
Query: 225 QIGDEILLPKKTKPKSPMSLSRPCKKLRT-GKSKDVGKSPELRKRKKVQVEGGEPSSLGP 283
+IG E +L K S S KK+ +SK K +R +V E + G
Sbjct: 24 KIGQETILSIDLKKSS--RASNTSKKMGLCPQSKAAHKKLAAPRRHRVDDELSSQAKAGL 197
Query: 284 HFT----AQGEARKYTPKNPSFTVSLKPV-IKLAYRPPVPISFIREYLNEKKQTIKLKIE 338
Q A+ +T PSF +K + +Y +P F +L K + L+
Sbjct: 198 RMLFALDEQRVAQAFTSPFPSFVKIMKKFNVSGSYTLKIPYQFSAAHLPTYKTEVTLRNS 377
Query: 339 EN-LWPVEFIYYPEHGSGK----LSKGWIQFAKESKLVGGDVCVFELINKDE 385
W V + G+ GW+ F +++ + GD C+FEL+ + E
Sbjct: 378 RGECWTVNSV---PDAKGRTVHTFCGGWMAFVRDNDINFGDTCIFELVAQCE 524
>AW781777
Length = 349
Score = 34.7 bits (78), Expect = 0.088
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +3
Query: 342 WPVEFIYYPEHGSGKLSKGWIQFAKESKLVGGDVCVFE 379
W + Y+ S L+KGW ++ K+ +L GDV +FE
Sbjct: 213 WRFRYSYWNSSQSYVLTKGWSRYVKDKRLDAGDVVLFE 326
Score = 33.5 bits (75), Expect = 0.20
Identities = 25/76 (32%), Positives = 35/76 (45%), Gaps = 12/76 (15%)
Frame = +3
Query: 127 LILPNGFMKKY------SGGMSNPVFLKAPDNTS---WEI---HWSKHDSYFWFQKGWKE 174
L++P +KY SGG L + ++ S W +W+ SY KGW
Sbjct: 102 LVIPKQHAEKYFPLSGDSGGSECKGLLLSFEDESGKCWRFRYSYWNSSQSYV-LTKGWSR 278
Query: 175 FVKYYSLDHGHLVLFE 190
+VK LD G +VLFE
Sbjct: 279 YVKDKRLDAGDVVLFE 326
>AI966696
Length = 333
Score = 30.8 bits (68), Expect = 1.3
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Frame = -2
Query: 146 FLKAPDNTSWEIHWSKHDS-YFWFQKGWKEFVKYYS 180
FLK N +W + H+ +F+F GWK F+ ++S
Sbjct: 236 FLKPDKNLTWILEEKIHERVFFFFLPGWKSFLFFFS 129
>TC209872
Length = 818
Score = 30.0 bits (66), Expect = 2.2
Identities = 20/58 (34%), Positives = 25/58 (42%), Gaps = 4/58 (6%)
Frame = +1
Query: 120 PCLEAAELILPNGFMKKYSGGMSN---PVFLKAPDNTSWE-IHWSKHDSYFWFQKGWK 173
PC E + L NG Y G S+ + N W IH+S +S FW GWK
Sbjct: 394 PCNELFDSCLCNGAGGVYGGCASSCGQQLCTFPSYNCHWSHIHFSHRESTFWIFSGWK 567
>TC223894
Length = 702
Score = 30.0 bits (66), Expect = 2.2
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Frame = +2
Query: 359 KGWIQFAKESKLVGGDVCVFELIN-KDEENPVLEVHI 394
KGW F + +K+ D C E+I+ +D+ L VH+
Sbjct: 443 KGWASFCRRNKIERNDTCFCEVISGEDQVVKTLRVHV 553
>CD396192
Length = 694
Score = 30.0 bits (66), Expect = 2.2
Identities = 12/22 (54%), Positives = 17/22 (76%)
Frame = +1
Query: 176 VKYYSLDHGHLVLFEFKDASHF 197
V+Y+SL HGHL+LF+ K +F
Sbjct: 88 VEYHSLSHGHLLLFKCKVTCNF 153
>CA802409 similar to GP|24198299|gb| Strictosidine synthase precursor
{Leptospira interrogans serovar lai str. 56601}, partial
(4%)
Length = 456
Score = 30.0 bits (66), Expect = 2.2
Identities = 15/44 (34%), Positives = 23/44 (52%)
Frame = -2
Query: 232 LPKKTKPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRKKVQVEG 275
+PK+ SP LS CK+ K + KSP ++R K+ + G
Sbjct: 389 IPKEVHKSSPC*LSPLCKQSMMNFHKSLRKSPRRKRRTKLGLSG 258
>CF921655
Length = 495
Score = 30.0 bits (66), Expect = 2.2
Identities = 24/85 (28%), Positives = 32/85 (37%), Gaps = 4/85 (4%)
Frame = +1
Query: 237 KPKSPMSLSRPCKKLRTGKSKDVGKSPELRKRK----KVQVEGGEPSSLGPHFTAQGEAR 292
+P P KK +T K G P+ K+K K + GGEP F
Sbjct: 7 RPPGPGEKGVKKKKKKTPAPKKWGPRPQYSKKKLAPKKKTLWGGEPKPQKGGFPLPLNWE 186
Query: 293 KYTPKNPSFTVSLKPVIKLAYRPPV 317
K PKNP + K+ PP+
Sbjct: 187 KKGPKNPGIPLGKNFNKKIPGNPPL 261
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,790,206
Number of Sequences: 63676
Number of extensions: 397090
Number of successful extensions: 1939
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 1893
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1935
length of query: 441
length of database: 12,639,632
effective HSP length: 100
effective length of query: 341
effective length of database: 6,272,032
effective search space: 2138762912
effective search space used: 2138762912
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0040.3


