

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
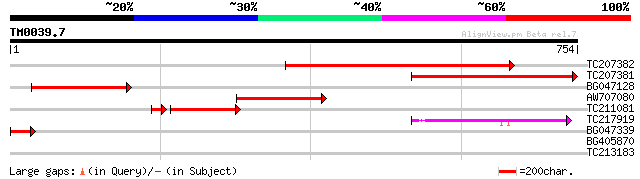
Query= TM0039.7
(754 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207382 similar to UP|Q8GUJ7 (Q8GUJ7) Dipeptidyl peptidase IV-l... 597 e-171
TC207381 similar to UP|Q8GUJ7 (Q8GUJ7) Dipeptidyl peptidase IV-l... 410 e-114
BG047128 similar to GP|10177085|dbj dipeptidyl peptidase IV-like... 235 5e-62
AW707080 similar to GP|10177085|dbj dipeptidyl peptidase IV-like... 217 2e-56
TC211081 similar to UP|Q8GUJ7 (Q8GUJ7) Dipeptidyl peptidase IV-l... 176 4e-45
TC217919 similar to UP|Q84LM4 (Q84LM4) Acylamino acid-releasing ... 62 9e-10
BG047339 62 9e-10
BG405870 30 3.0
TC213183 similar to UP|Q6YUW7 (Q6YUW7) Chromosome structural mai... 30 3.0
>TC207382 similar to UP|Q8GUJ7 (Q8GUJ7) Dipeptidyl peptidase IV-like protein
(At5g24260), partial (53%)
Length = 915
Score = 597 bits (1539), Expect = e-171
Identities = 280/304 (92%), Positives = 293/304 (96%)
Frame = +2
Query: 368 TGFRHLYLHDANGTCLGPITEGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLYCAKLYV 427
TGFRHLYLHDANGTCLGPITEGEWMVEQIAGVNEATGL+YFTGTLDGPLESNLYCAKL+V
Sbjct: 2 TGFRHLYLHDANGTCLGPITEGEWMVEQIAGVNEATGLIYFTGTLDGPLESNLYCAKLFV 181
Query: 428 DGTHPPEAPTRLTLNKGKHIVVLDHHMQSFVDIYDSLGCPPRVLLCSLEDGRVIMPLFEQ 487
DG P +AP RLT +KGKHIVVLDHHMQSFVDI+DSLGCPPRVLLCSLEDG VI PL+EQ
Sbjct: 182 DGNQPLQAPIRLTHSKGKHIVVLDHHMQSFVDIHDSLGCPPRVLLCSLEDGSVIKPLYEQ 361
Query: 488 PFSVPRFKKLQLEPPEIVEIQANDGTVLYGALYKPDASRFGPPPYKTMINVYGGPSVQLV 547
F++PRFKKLQLEPPEIVEIQANDGT LYGALYKPDAS+FGPPPYKTMINVYGGPSVQLV
Sbjct: 362 SFTIPRFKKLQLEPPEIVEIQANDGTTLYGALYKPDASKFGPPPYKTMINVYGGPSVQLV 541
Query: 548 SNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQVDADDQLTGAEWL 607
NSWL+TVD+RAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQ+DADDQLTGAEWL
Sbjct: 542 CNSWLSTVDLRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQIDADDQLTGAEWL 721
Query: 608 VKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWDGYDTFYTEKYMGL 667
VKQGLAK GHIGLYGWSYGGYLSAMTLSRYPDFFKCA+AGAPVTSWDGYDTFYTEKYMGL
Sbjct: 722 VKQGLAKGGHIGLYGWSYGGYLSAMTLSRYPDFFKCAIAGAPVTSWDGYDTFYTEKYMGL 901
Query: 668 PSEN 671
PSEN
Sbjct: 902 PSEN 913
>TC207381 similar to UP|Q8GUJ7 (Q8GUJ7) Dipeptidyl peptidase IV-like protein
(At5g24260), partial (36%)
Length = 841
Score = 410 bits (1054), Expect = e-114
Identities = 196/220 (89%), Positives = 205/220 (93%)
Frame = +2
Query: 535 MINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQ 594
MINVYGGPSVQLVSNS L+TVD+R RNQGILVWKL NRGTA GLK ESYLK KLGQ
Sbjct: 2 MINVYGGPSVQLVSNSXLSTVDLRXXXXRNQGILVWKLXNRGTAXXGLKIESYLKQKLGQ 181
Query: 595 VDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTSWD 654
+DADDQLTGAEWLVKQGL KAGHIGLYGWSYGGYLSAMTLSRYPDFFKCA+AGAPVTSWD
Sbjct: 182 IDADDQLTGAEWLVKQGLTKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAIAGAPVTSWD 361
Query: 655 GYDTFYTEKYMGLPSENQSEYESGSVMNQVHKLKGRLLLVHGMIDENVHFRHTARLINAL 714
GYDTFYTEKYMGLPSEN+S YESGSVMNQVH+LKGRLLLVHGMIDENVHFRHTARLINAL
Sbjct: 362 GYDTFYTEKYMGLPSENKSGYESGSVMNQVHQLKGRLLLVHGMIDENVHFRHTARLINAL 541
Query: 715 VAAGKPYELVLFPDERHMPRRHSDRVYMEERMWEFIDRNL 754
VAAGK YEL++FPDERHMPRRHSDRVYME RMW+FI RNL
Sbjct: 542 VAAGKSYELIVFPDERHMPRRHSDRVYMEGRMWDFIQRNL 661
>BG047128 similar to GP|10177085|dbj dipeptidyl peptidase IV-like protein
{Arabidopsis thaliana}, partial (13%)
Length = 399
Score = 235 bits (600), Expect = 5e-62
Identities = 116/133 (87%), Positives = 125/133 (93%)
Frame = +1
Query: 30 GYVSPSSINFSPDDSLISYLYSPEQTLNRKVFAFNVKTKAQELLFSPPDGGLDESNISPE 89
GYVSP+SI+FSPDDSLISYL+SP+ +LNRKV+AF++KT QELLFSPPDGGLDESNISPE
Sbjct: 1 GYVSPTSISFSPDDSLISYLFSPDNSLNRKVYAFDLKTNTQELLFSPPDGGLDESNISPE 180
Query: 90 EKLRRERLRERGLGVTRYEWVKTSSKRKAVMVPLPAGIYIQDISQSKAELKLSSIPSSPI 149
EKLRRERLRERGLGVTRYEWVKTSSKRKAVMVPLPAGIYIQDI SKAELKL S+ SPI
Sbjct: 181 EKLRRERLRERGLGVTRYEWVKTSSKRKAVMVPLPAGIYIQDIPHSKAELKLPSVLGSPI 360
Query: 150 IDPHLSPDGSMLA 162
IDPHLSPDGSMLA
Sbjct: 361 IDPHLSPDGSMLA 399
>AW707080 similar to GP|10177085|dbj dipeptidyl peptidase IV-like protein
{Arabidopsis thaliana}, partial (9%)
Length = 367
Score = 217 bits (552), Expect = 2e-56
Identities = 100/120 (83%), Positives = 109/120 (90%)
Frame = +2
Query: 302 GNTLTAQVLNRHHTKIKILKFDIRTGQGKNILVEENSTWINIHDCFTPLDKGITKFSGGF 361
GN LTAQ+LNRHHTKIKI+KFDIRTGQ KNIL ++N WINIHDCFTPLDKG+TKFSGGF
Sbjct: 5 GNILTAQILNRHHTKIKIVKFDIRTGQKKNILFKKNXNWINIHDCFTPLDKGVTKFSGGF 184
Query: 362 IWASEKTGFRHLYLHDANGTCLGPITEGEWMVEQIAGVNEATGLLYFTGTLDGPLESNLY 421
IWA+EKT F+HLYLHDANGTCL PITE EWMVEQIA VNEAT L+YFT TLDGPL+SNLY
Sbjct: 185 IWANEKTRFKHLYLHDANGTCLRPITESEWMVEQIASVNEATCLIYFTETLDGPLKSNLY 364
>TC211081 similar to UP|Q8GUJ7 (Q8GUJ7) Dipeptidyl peptidase IV-like protein
(At5g24260), partial (10%)
Length = 367
Score = 176 bits (447), Expect(2) = 4e-45
Identities = 86/93 (92%), Positives = 88/93 (94%)
Frame = +3
Query: 215 DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHPYPFAGASNVKVRLGVVSAAGSSI 274
DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDH YPFAGASNVKVRLGVVS AGSSI
Sbjct: 87 DSKYIAFTEVDSSEIPLFRIMHQGKSSVGLEAQEDHSYPFAGASNVKVRLGVVSVAGSSI 266
Query: 275 TWMDLLCGGKEQQANEEEYLARVNWMHGNTLTA 307
TWM+LLCGG EQQ N+EEYLARVNWMHGN LTA
Sbjct: 267 TWMNLLCGGTEQQNNDEEYLARVNWMHGNILTA 365
Score = 24.3 bits (51), Expect(2) = 4e-45
Identities = 13/21 (61%), Positives = 14/21 (65%), Gaps = 1/21 (4%)
Frame = +1
Query: 189 NGLTRGLAEYIAQ-EEMDRKN 208
NGLT GLA YIA DR+N
Sbjct: 1 NGLTHGLA*YIAS*ARCDRRN 63
>TC217919 similar to UP|Q84LM4 (Q84LM4) Acylamino acid-releasing enzyme ,
partial (33%)
Length = 1228
Score = 62.0 bits (149), Expect = 9e-10
Identities = 60/231 (25%), Positives = 105/231 (44%), Gaps = 18/231 (7%)
Frame = +2
Query: 535 MINVYGGPSVQLVSNSWLNTVDMRAQYLRNQGILVWKLDNRGTARRGLKFESYLKHKLGQ 594
++ ++GGP VS L++ YL + G + ++ RG+ G + L K G
Sbjct: 272 IVVLHGGPHT--VS---LSSFSKPLAYLSSLGYSLLIVNYRGSLGFGEEALQSLPGKAGS 436
Query: 595 VDADDQLTGAEWLVKQGLAKAGHIGLYGWSYGGYLSAMTLSRYPDFFKCAVAGAPVTS-- 652
D +D LT + ++ GLA I + G S+GG+L+ + + P+ F A A PV +
Sbjct: 437 QDVNDVLTAIDHVINLGLASPSKIAVLGGSHGGFLTTHLIGQAPEKFVAAAARNPVCNLA 616
Query: 653 ----------WDGYDTFYT---EKYMGLPS-ENQSEYESGSVMNQVHKLKGRLLLVHGMI 698
W +T+ T +K+ PS E+ + + S S + + K+K + + G
Sbjct: 617 LMVGTTDIPDWCYVETYGTKGRDKFTEAPSAEDLTLFYSKSPIAHLSKVKTPTIFLLGAQ 796
Query: 699 DENVHFRHTARLINALVAAGKPYELVLFPDERH-MPRRHSD-RVYMEERMW 747
D V + AL G ++++FP++ H + R SD Y+ MW
Sbjct: 797 DLRVPISTGLQYARALREKGGQVKVIVFPNDVHGIERPQSDFESYLNIAMW 949
>BG047339
Length = 422
Score = 62.0 bits (149), Expect = 9e-10
Identities = 28/34 (82%), Positives = 30/34 (87%)
Frame = +3
Query: 1 MPVTDFSDVPNIDDNILFPVEEIVQYPLPGYVSP 34
MPVTD ++ N DDNILFPVEEIVQYPLPGYVSP
Sbjct: 321 MPVTDSNNPQNFDDNILFPVEEIVQYPLPGYVSP 422
>BG405870
Length = 307
Score = 30.4 bits (67), Expect = 3.0
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Frame = -1
Query: 692 LLVHGMIDENVHFRHTA-RLINALVAAGKPYELVLFPDERHMPRRHSDRV 740
L+ H D FR R ++ +++G+P+E LFP E H P+ S +V
Sbjct: 262 LMTHN**DSQKVFRAIEQRQLDLQLSSGEPWEKQLFPQE*HTPQHQSSQV 113
>TC213183 similar to UP|Q6YUW7 (Q6YUW7) Chromosome structural maintenance
protein-like, partial (3%)
Length = 517
Score = 30.4 bits (67), Expect = 3.0
Identities = 20/69 (28%), Positives = 34/69 (48%)
Frame = +3
Query: 15 NILFPVEEIVQYPLPGYVSPSSINFSPDDSLISYLYSPEQTLNRKVFAFNVKTKAQELLF 74
N++FP+E + P P ++SPS+++F L L++ F ++T+ EL
Sbjct: 99 NLIFPMEWLQTLPPPSHLSPSALSF-----LDHRLHTQLALAEAPTFVAELQTQCSEL-- 257
Query: 75 SPPDGGLDE 83
D LDE
Sbjct: 258 ---DRSLDE 275
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,145,138
Number of Sequences: 63676
Number of extensions: 529349
Number of successful extensions: 2159
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 2141
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2158
length of query: 754
length of database: 12,639,632
effective HSP length: 104
effective length of query: 650
effective length of database: 6,017,328
effective search space: 3911263200
effective search space used: 3911263200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Lotus: description of TM0039.7


