

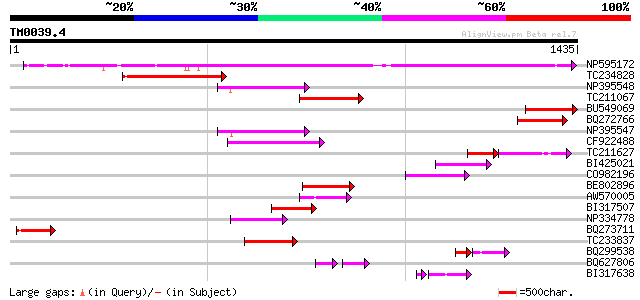
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.4
(1435 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NP595172 polyprotein [Glycine max] 740 0.0
TC234828 274 2e-73
NP395548 reverse transcriptase [Glycine max] 168 2e-41
TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%) 161 2e-39
BU549069 159 6e-39
BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis ... 155 1e-37
NP395547 reverse transcriptase [Glycine max] 154 4e-37
CF922488 140 3e-33
TC211627 79 4e-31
BI425021 117 3e-26
CO982196 114 2e-25
BE802896 114 4e-25
AW570005 105 1e-22
BI317507 102 9e-22
NP334778 reverse transcriptase [Glycine max] 102 2e-21
BQ273711 96 1e-19
TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, part... 94 6e-19
BQ299538 59 4e-18
BQ627806 58 7e-16
BI317638 weakly similar to GP|9294238|dbj| contains similarity t... 78 4e-15
>NP595172 polyprotein [Glycine max]
Length = 4659
Score = 740 bits (1911), Expect = 0.0
Identities = 485/1451 (33%), Positives = 730/1451 (49%), Gaps = 52/1451 (3%)
Frame = +1
Query: 35 LKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYASYLFHEEARNWWVHAKSR 94
+K + P FDG + W+ K EQ ++ T + + AS ++ W+
Sbjct: 292 VKLDFPRFDG----KNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWY----QM 447
Query: 95 ITPPDGVLTWSIFKEAFLEKYFPADVKGKKETEFLELKQGEMFVGQYAARFEELSQFHPY 154
+ + +W F A + P+ + T F +L Q V +Y +F L +
Sbjct: 448 LQKTEPFSSWQAFTRALELDFGPSAYDCPRATLF-KLNQSAT-VNEYYMQFTAL--VNRV 615
Query: 155 YGTTADDASKCIRFECGLRPDIRAAIGHQQIRTFTVLVEKCRIFEENDRARREYYKSSKF 214
G +A+ C F GL+ +I + + RT T V ++FEE + + S
Sbjct: 616 DGLSAEAILDC--FVSGLQEEISRDVKAMEPRTLTKAVALAKLFEEKYTSPPKTKTFSNL 789
Query: 215 NKTSKRREERKKPYSPRNYKPE-------------------LQNRNYGGARPTN---PNS 252
+ + Y P N K + L+N+N P
Sbjct: 790 ARNFTSNTSATQKYPPTNQKNDNPKPNLPPLLPTPSTKPFNLRNQNIKKISPAEIQLRRE 969
Query: 253 HVTCYRCGKEGHKSWSCTATTTSGQTNLKPPVVGGTNTTSAPRNSAGASAQKPGRPVNKG 312
CY C ++ + C + ++ T + + +
Sbjct: 970 KNLCYFCDEKFSPAHKCPN---------RQVMLLQLEETDEDQTDEQVMVTEEANMDDDT 1122
Query: 313 KVFAMTGAEANTSEDFIQGTCFLCDISLVVLYDSGATHSFISHERAKSLKLVITQLPYDL 372
++ + I+ T + I++ +L D G++ +FI A+ LKL + P +L
Sbjct: 1123 HHLSLNAMRGSNGVGTIRFTGQVGGIAVKILVDGGSSDNFIQPRVAQVLKLPVEPAP-NL 1299
Query: 373 VVTTPTKESAVTSSVCKKCPLVIEDREYITNLVCLPLEGLDIILGMNWL-SINNVLLDCR 431
V + + ++ PL I+ +E + L + G D+ILG WL ++ + D
Sbjct: 1300 RVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVILGSTWLATLGPHVADYA 1479
Query: 432 LRVPIFLQKYK----------EKHTASL--------PEKEPSAYLILFSSEGTKRPAMED 473
F Q K E A L + + I + ++D
Sbjct: 1480 ALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECFAIQLIQKEVPEDTLKD 1659
Query: 474 IP---------VVREFPEVFPEDMTELPPEREVEFAIDVIPGTTPISAAPYRISPLELAE 524
+P ++ + +VF LPP+RE + AI + G+ P+ PYR + +
Sbjct: 1660 LPTNIDPELAILLHTYAQVFAVP-ASLPPQREQDHAIPLKQGSGPVKVRPYRYPHTQKDQ 1836
Query: 525 LQKQVEELLSKGFIRPSVSPWGAPVLLVKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDD 584
++K ++E+L +G I+PS SP+ P+LLVKKKDGS R C DYR LN +T+K+ +P+P +D+
Sbjct: 1837 IEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDYRALNAITVKDSFPMPTVDE 2016
Query: 585 LMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMD 644
L+D+L GA+ FSK+DLRSGYHQI V+ +D +KTAFRT +GHYE+LVMPFG+TNAPA F
Sbjct: 2017 LLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGHYEWLVMPFGLTNAPATFQC 2196
Query: 645 YMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQ 704
MN+IF L KFV+VF DDILIYS S ++H++H++ VL+ LK +L+A+LSKC F +
Sbjct: 2197 LMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQTLKQHQLFARLSKCSFGDTE 2376
Query: 705 VQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIATPLT 764
V +LGH VS G++++ KV+AV W P V +R FLGL GYYRRFI+ ++ IA PLT
Sbjct: 2377 VDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGLTGYYRRFIKSYANIAGPLT 2556
Query: 765 QLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQER 824
L +KD F+W + E +F LK+ +T+APVL+LPD S+ + + DAS G+G VL Q
Sbjct: 2557 DLLQKD-SFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFILETDASGIGVGAVLGQNG 2733
Query: 825 KVIAYASQQLRPHEQNYPTHDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKT 884
IAY S++L P Q + EL A+ AL +RHYL G KF I +D +SLK L DQ
Sbjct: 2734 HPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNKFIIRTDQRSLKSLMDQSL 2913
Query: 885 LNMRQRRWVEFLEDYDFKLQYHPGKANVVADALSRKSLHAARLMIEETELIEKFRDMNLI 944
Q+ W+ YDFK++Y PGK N ADALSR M ++
Sbjct: 2914 QTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSR---------------------MFML 3030
Query: 945 METLPQGTRLGTLTLTNEFIEEVKKEQARDENLQKEAHGRDSMSRPDFLKGPDGLWRYQG 1004
+ P F+EE++ D +L++ + +GL ++
Sbjct: 3031 AWSEPHSI----------FLEELRARLISDPHLKQLMETYKQGADASHYTVREGLLYWKD 3180
Query: 1005 RLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCLTCQ 1064
R+ +P E+ KIL+E H S H G T+ LK F+WP M++D+ + CL CQ
Sbjct: 3181 RVVIPAEEEIVNKILQEYHSSPIGGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQ 3360
Query: 1065 KVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAV 1124
+ K + P+G LQPL IP+ WE ++MDF++GLP + G I V++DRLTK AHFI +
Sbjct: 3361 QAKSNNTLPAGLLQPLPIPQQVWEDVAMDFITGLP-NSFGLSVIMVVIDRLTKYAHFIPL 3537
Query: 1125 NMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQ 1184
+ S+ +A ++ IV+LHG+P +IVSDRD F S FW L + GT L++SSAYHPQ
Sbjct: 3538 KADYNSKVVAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQ 3717
Query: 1185 SDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLGMAPFEALYGRRCK 1244
SDGQSE + LE LR ++ W LP AEF YN +YH SLGM PF ALYGR
Sbjct: 3718 SDGQSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPP 3897
Query: 1245 TPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKRHKPLEFQEGDHVF 1304
T D E L + + ++ L AQ K DK+ + FQ GD V
Sbjct: 3898 TLTRQACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVL 4077
Query: 1305 LRVTPITGVGRSIH-SKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRK 1363
+++ P + ++KL+ +Y GP+++L +IG VAY++ L PS + +H VFH+SQL+
Sbjct: 4078 VKLQPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLEL-PSAARIHPVFHVSQLKP 4254
Query: 1364 YLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAWSDDNQD-ATWE 1422
+ + P + + E + QPVKIL R + + + W + QD ATWE
Sbjct: 4255 FNGTAQDPYLPLPLTVTE-MGPVMQPVKILASRIIIRGHNQIEQILVQWENGLQDEATWE 4431
Query: 1423 LEESARKRYPS 1433
E + YP+
Sbjct: 4432 DIEDIKASYPT 4464
>TC234828
Length = 857
Score = 274 bits (700), Expect = 2e-73
Identities = 139/268 (51%), Positives = 185/268 (68%), Gaps = 5/268 (1%)
Frame = +3
Query: 286 GGTNTTSAPRNSAGASAQKPGRPVNKGKVFAMTGAEANTSEDFIQGTCFLCDISLVVLYD 345
G TN+T + + + GRP +VFAM+G+EA S+D I+G C + D L VLYD
Sbjct: 57 GPTNSTRS--GNVSNNNISGGRPKVPSRVFAMSGSEAAASDDLIRGKCLIADKLLDVLYD 230
Query: 346 SGATHSFISHERAKSLKLVITQLPYDLVVTTPTKESAVTSSVCKKCPLVIEDREYITNLV 405
SGATHSFISH + L L T+LPYD+VV+TPT E TS VC KCP+++E R ++ +L+
Sbjct: 231 SGATHSFISHACVERLGLCATELPYDMVVSTPTSEPVTTSRVCLKCPIIVEGRSFMADLI 410
Query: 406 CLPLEGLDIILGMNWLSINNVLLDCRLRVPIF----LQKYKEKHTASLPEKEP-SAYLIL 460
CLPL LD+ILGM+WLS N++ LDC+ ++ +F + K A+ E E Y++L
Sbjct: 411 CLPLAHLDVILGMDWLSTNHIFLDCKEKMLVFGGDVVPSEPLKEDAANEETEDVRTYMVL 590
Query: 461 FSSEGTKRPAMEDIPVVREFPEVFPEDMTELPPEREVEFAIDVIPGTTPISAAPYRISPL 520
FS + + IPVV EFPEVFP+D+ ELPPEREVEF IDV+PG P+S APYR+SP+
Sbjct: 591 FSMYVEEDAEVSCIPVVSEFPEVFPDDVCELPPEREVEFIIDVVPGANPVSIAPYRMSPV 770
Query: 521 ELAELQKQVEELLSKGFIRPSVSPWGAP 548
ELAE++ QV++LLSK F+RPS SPWGAP
Sbjct: 771 ELAEVKAQVQDLLSKQFVRPSASPWGAP 854
>NP395548 reverse transcriptase [Glycine max]
Length = 762
Score = 168 bits (425), Expect = 2e-41
Identities = 93/254 (36%), Positives = 143/254 (55%), Gaps = 19/254 (7%)
Frame = +1
Query: 525 LQKQVEELLSKGFIRP-SVSPWGAPVLLVKKKDG------------------SMRLCVDY 565
++K+V +LL G I P S S W +PVL+V KK+G S +LC+DY
Sbjct: 1 VRKEVLKLLEVGLIYPISDSAWVSPVLVVSKKEGMTVIRNEKNDLIPTRTVTSWKLCIDY 180
Query: 566 RQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGH 625
R+LN+ T K+ +PLP +D ++++L G + +D GY+QI V D +K AF +G
Sbjct: 181 RKLNEATRKDHFPLPFMDQMLERLAGHAYYCFLDAYFGYNQIVVDPKDQEKMAFTCPFGV 360
Query: 626 YEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKV 685
+ Y +PFG+ NAP F M IF ++K + VF+DD ++ S E ++ +++VL+
Sbjct: 361 FAYRRIPFGLCNAPTTFQMCMLAIFADIVEKSIEVFMDDFSVFVPSLESCLKKLEMVLQR 540
Query: 686 LKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGL 745
+ L KC F + + LGH +S GI VD K++ + P V G+RSFLG
Sbjct: 541 CVETNLVLNWEKCHFMVREGIVLGHKISTRGIEVDQTKIDVIEKLPPPSNVKGIRSFLGQ 720
Query: 746 AGYYRRFIEGFSKI 759
A +YRRFI+ F+K+
Sbjct: 721 ARFYRRFIKDFTKV 762
>TC211067 similar to UP|Q9LQH2 (Q9LQH2) F15O4.13, partial (9%)
Length = 589
Score = 161 bits (408), Expect = 2e-39
Identities = 81/161 (50%), Positives = 109/161 (67%)
Frame = +1
Query: 734 ETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKA 793
++V +RSF GLA +YRRF+ FS +A+PL +L KK+ F W EK E +F LKE+LTKA
Sbjct: 106 KSVGDIRSFHGLASFYRRFVPNFSTVASPLNELVKKNMAFTWGEKQEQAFALLKEKLTKA 285
Query: 794 PVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVVF 853
PVL LPD SK +++ CDAS G+ VL+Q IAY S++L NYPT+D EL A++
Sbjct: 286 PVLALPDFSKTFELECDASGVGVRAVLLQGGHPIAYFSEKLHSATLNYPTYDKELYALIR 465
Query: 854 ALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVE 894
A + W H+L +F I+SDHQSLKY+ + LN R +WVE
Sbjct: 466 APQTWEHFLVCKEFVIHSDHQSLKYIRGKSKLNKRHAKWVE 588
>BU549069
Length = 615
Score = 159 bits (403), Expect = 6e-39
Identities = 76/132 (57%), Positives = 102/132 (76%), Gaps = 1/132 (0%)
Frame = -1
Query: 1305 LRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFHISQLRKY 1364
L+VTP TGVG+++ S+KLTP ++G +QIL R VAY+IALPPSLSNLH+VFH+SQLR Y
Sbjct: 615 LKVTPRTGVGQALKSRKLTPHFIGHFQILKRAXPVAYQIALPPSLSNLHNVFHVSQLRMY 436
Query: 1365 LPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTVPLVKLAW-SDDNQDATWEL 1423
+ D SHV++ D+++++ENLTY T P++I +RR K LR++ PLVK+ W +DATWEL
Sbjct: 435 IHDPSHVVKLDDVQVKENLTYETLPLRIEDRRTKHLRRKENPLVKVIWGGTSGEDATWEL 256
Query: 1424 EESARKRYPSLF 1435
E R YPSLF
Sbjct: 255 ESQMRVAYPSLF 220
>BQ272766 weakly similar to GP|28558781|gb| pol protein {Cucumis melo}, partial
(9%)
Length = 410
Score = 155 bits (392), Expect = 1e-37
Identities = 71/127 (55%), Positives = 100/127 (77%)
Frame = -3
Query: 1286 SYYDKRHKPLEFQEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIAL 1345
SY+DKR K LEF+ GDHVFLRVTP TGVGR++ S KLTP ++GP+QIL ++ VAY+IAL
Sbjct: 408 SYHDKRRKDLEFEVGDHVFLRVTP*TGVGRALKS*KLTPHFIGPFQILKKVDFVAYQIAL 229
Query: 1346 PPSLSNLHDVFHISQLRKYLPDSSHVIEPDNIELEENLTYPTQPVKILERREKQLRKRTV 1405
PPSL++LH+VFH+SQ+ KY+ D SH++ D ++++ENL + T P++I + R K LR + +
Sbjct: 228 PPSLTSLHNVFHVSQIHKYINDPSHMVNLDVVQVKENLAHETLPLRIKDMRTKHLRGKEI 49
Query: 1406 PLVKLAW 1412
LVK+ W
Sbjct: 48 LLVKVIW 28
>NP395547 reverse transcriptase [Glycine max]
Length = 762
Score = 154 bits (388), Expect = 4e-37
Identities = 89/254 (35%), Positives = 136/254 (53%), Gaps = 19/254 (7%)
Frame = +1
Query: 525 LQKQVEELLSKGFIRP-SVSPWGAPVLLVKKKDGSM------------------RLCVDY 565
++K+V +LL G I P S S W +PV +V KK G R+C+DY
Sbjct: 1 VRKEVFKLLEAGLIYPISDSSWVSPVQVVPKKGGMTVVKNDRNELIPTRRVTRWRMCIDY 180
Query: 566 RQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAFRTRYGH 625
R+LN+ T K+ YPLP +D ++ +L + +D SGY+QI V D +KTAF +
Sbjct: 181 RKLNEATRKDHYPLPFMDQMLKRLARQSFYRFLDGYSGYNQIAVDPQDQEKTAFTCPFSV 360
Query: 626 YEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHMQVVLKV 685
+ Y MPFG+ NA F M IF ++K + VF+DD + S + +++ VL+
Sbjct: 361 FAYRRMPFGLCNASTTFQRCMMAIFDDMVEKCIEVFMDDFSFFGASFGNCLANLEKVLQR 540
Query: 686 LKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWKVPETVTGVRSFLGL 745
+ L KC F +++ LGH +S+ GI V K++ ++ P V G+ SFLG
Sbjct: 541 CEKSNLVLNWEKCHFMVQEGIVLGHKISKRGIEVVKEKLDVIDKLPPPVNVKGIHSFLGH 720
Query: 746 AGYYRRFIEGFSKI 759
G+YRRFI+ F+K+
Sbjct: 721 VGFYRRFIKDFTKV 762
>CF922488
Length = 741
Score = 140 bits (354), Expect = 3e-33
Identities = 82/245 (33%), Positives = 134/245 (54%)
Frame = +3
Query: 552 VKKKDGSMRLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKS 611
V K+DG + +CVDYR LN + K+++PLP I+ L+D FS +D SGY+QI++
Sbjct: 3 VLKEDGKV*MCVDYRDLN*ASPKDKFPLPHINVLVDNTTSFSQFSFMDGFSGYNQIKIAP 182
Query: 612 DDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKS 671
+D++KT F T +G + Y M FG+ N A + M +F + K + V++DD+++ S++
Sbjct: 183 EDMEKTTFITLWGTFCYKAMSFGLKNVGATYQRAMVALF*DMMHKEIEVYMDDMIVKSRT 362
Query: 672 KEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAKVEAVNSWK 731
+EEH+ +++ + + L+ +L +KC F ++ + L + S GI VD KV+ +
Sbjct: 363 EEEHLVNLRKLFRRLRKYRLRLNPAKCMFEVKSRKLLDFIDS*RGIEVDSNKVKVILEMA 542
Query: 732 VPETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLT 791
P T V+ FLG Y RFI PL L K+ W C +F+ +K+ L
Sbjct: 543 KPHTEKQVQGFLGRLNYIVRFIS*LIATCEPLFILLCKNQFVKWDHDC*VAFERIKQCLI 722
Query: 792 KAPVL 796
VL
Sbjct: 723 NPHVL 737
>TC211627
Length = 1034
Score = 78.6 bits (192), Expect(2) = 4e-31
Identities = 39/78 (50%), Positives = 48/78 (61%)
Frame = +2
Query: 1159 FVSKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLA 1218
F+S W L GT+L S+AYHPQ+DGQ+E + LE LRA V D+ W FL LA
Sbjct: 5 FISGLWHELFHISGTKLRFSTAYHPQTDGQTEVINRILEQYLRAFVHDHPQHWFKFLSLA 184
Query: 1219 EFSYNNSYHSSLGMAPFE 1236
E YN S HS +G +PFE
Sbjct: 185 E*CYNTSVHSGIGFSPFE 238
Score = 76.3 bits (186), Expect(2) = 4e-31
Identities = 52/189 (27%), Positives = 91/189 (47%), Gaps = 4/189 (2%)
Frame = +3
Query: 1238 LYGRRCKTPLCWLSGEDKITLGPELLQEMTEKVRSIREKLRIAQDRQKSYYDKRHKPLEF 1297
+YG+ + +G + +L + ++ +L+ QD K D + L F
Sbjct: 243 MYGKPPPALPLYSAGTSTVEAVDAILHSLATIHHTLTCRLQKYQDSMKRIADSHRRDLTF 422
Query: 1298 QEGDHVFLRVTPITGVGRSIHSKKLTPKYLGPYQILDRIGAVAYRIALPPSLSNLHDVFH 1357
GD V++R+ P KL+ ++ GPYQI R+G VAYR+ LPP+ S +H +FH
Sbjct: 423 NIGDWVYVRL*PYRQTSIQSTYTKLSKRFYGPYQIQARVGQVAYRLQLPPT-SKIHPIFH 599
Query: 1358 ISQLRKYLPDSSHVIEPDNIELE--ENLTYP-TQPVKILERREKQLRKRTVPLVKLAWSD 1414
+S L+ + I P+ + L +P QP++ L+ + + +P V + W++
Sbjct: 600 VSLLKVH----HGPIPPELLALPPFSTTNHPLVQPLQFLDWKMDESTTPPIPQVLVQWTN 767
Query: 1415 -DNQDATWE 1422
+D TWE
Sbjct: 768 LAPEDTTWE 794
>BI425021
Length = 426
Score = 117 bits (294), Expect = 3e-26
Identities = 60/143 (41%), Positives = 87/143 (59%)
Frame = -1
Query: 1077 LQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHFIAVNMTFPSEKLARI 1136
L PL +P+ WE +SMDF+ GLP GH I+V+V+R +K H + + + +A +
Sbjct: 426 LCPLPVPQRPWEDLSMDFIVGLP-PYHGHTTIFVVVNRFSKGIHLGTLPTSHTAHMVASL 250
Query: 1137 YVKEIVRLHGVPANIVSDRDPRFVSKFWGSLHEALGTRLSLSSAYHPQSDGQSERTIQTL 1196
++ +++LHG P +IVSDRDP F+S FW L GT L +SSAYHPQ+DGQ+E + +
Sbjct: 249 FLNIVIKLHGFPRSIVSDRDPLFISHFWQDLFRLSGTVLRMSSAYHPQTDGQTEVLNRVI 70
Query: 1197 EDMLRACVLDYKGSWEDFLPLAE 1219
E LRA V + F+P E
Sbjct: 69 EQYLRAFVHGRPRNLGRFIPWVE 1
>CO982196
Length = 812
Score = 114 bits (286), Expect = 2e-25
Identities = 62/162 (38%), Positives = 94/162 (57%)
Frame = +1
Query: 1002 YQGRLCVPEGGELRQKILEEGHKSDFSIHPGTTKMYQDLKKMFWWPGMKKDIMKKVTSCL 1061
++ RL + + +L+E S H G + ++ + + +W GMKK V +C
Sbjct: 328 FKDRLVLSKNSTKIPLLLKELQDSPLGGHSGFFRTFKRVANVVFWQGMKKTTRDYVAACE 507
Query: 1062 TCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKSAHF 1121
C++ K P+G L L IP W ISMDF+ GLP+ G D I V+VDRLTK AHF
Sbjct: 508 ICRRNKTSTLSPAGLL*LLPIPTKVWTDISMDFIGGLPKAQ-GKDNILVVVDRLTKYAHF 684
Query: 1122 IAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKF 1163
A++ + ++++A +++KE+VRLHG PA+IVSD F+S F
Sbjct: 685 FALSHPYTAKEVAELFIKELVRLHGFPASIVSDXXRLFMSLF 810
Score = 30.0 bits (66), Expect = 7.7
Identities = 13/34 (38%), Positives = 22/34 (64%)
Frame = +2
Query: 859 RHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRW 892
RHY G KF I ++ +S K+L +Q+ ++ Q +W
Sbjct: 53 RHYPVGKKFIIRTN*RSSKFLNEQRLMSEEQFKW 154
>BE802896
Length = 416
Score = 114 bits (284), Expect = 4e-25
Identities = 59/137 (43%), Positives = 85/137 (61%), Gaps = 4/137 (2%)
Frame = -2
Query: 741 SFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTKAPVLTLPD 800
SFLG AG+YRRFI F K+A PL+ L +K+ F + +KC+*+F LK L P++ PD
Sbjct: 415 SFLGHAGFYRRFIRDFRKVALPLSNLLQKEVEFDFNDKCK*AFDCLKRALITTPIIQAPD 236
Query: 801 PSKDYDVYCDASKSGLGCVLMQE----RKVIAYASQQLRPHEQNYPTHDMELAAVVFALK 856
+ +++ CDAS LG VL Q+ +VI Y+S+ L + NY T + EL A+VFAL+
Sbjct: 235 WTAPFELMCDASNYALGVVLAQKIDKLPRVIYYSSRTLDAAQANYTTTEKELLAIVFALE 56
Query: 857 IWRHYLYGVKFTIYSDH 873
+ YL G + +Y DH
Sbjct: 55 KFHSYLLGTRIIVYIDH 5
>AW570005
Length = 413
Score = 105 bits (262), Expect = 1e-22
Identities = 58/132 (43%), Positives = 79/132 (58%)
Frame = -2
Query: 733 PETVTGVRSFLGLAGYYRRFIEGFSKIATPLTQLTKKDHPFVWTEKCE*SFQTLKERLTK 792
P T +R FL L G+YRRFI+G++ +A PL+ L KD FVW+ + + +FQ LK +T
Sbjct: 406 PRTARSLRGFLRLTGFYRRFIKGYAAMAAPLSHLLTKDS-FVWSPEADVAFQALKNVVTN 230
Query: 793 APVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAYASQQLRPHEQNYPTHDMELAAVV 852
VL LPD +K + V DAS S +G VL QE IA+ S++ P T+ ELAA+
Sbjct: 229 TLVLALPDFTKPFTVETDASGSDMGAVLSQEGHPIAFFSKEFCPKLVRSSTYVHELAAIT 50
Query: 853 FALKIWRHYLYG 864
+K WR YL G
Sbjct: 49 NVVKKWRQYLLG 14
>BI317507
Length = 359
Score = 102 bits (255), Expect = 9e-22
Identities = 54/113 (47%), Positives = 73/113 (63%), Gaps = 1/113 (0%)
Frame = -1
Query: 664 DILIYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSEDGIAVDPAK 723
+ILIYS + H+ H+ VL VLK +L A KC F +++LGHV+S+D +A+D K
Sbjct: 353 NILIYSPDWKSHIMHLTAVLDVLKKERLVANRKKCYFSQTTIEYLGHVISKDCVAMDSNK 174
Query: 724 VEAVNSWKVPETVTGVRSFLGLAGYYRRFIEGFSKIA-TPLTQLTKKDHPFVW 775
V++V W VP+ V V SFL L GYYR+FI+ + K+A PLT LTK D F W
Sbjct: 173 VKSVIEWPVPKNVKRVCSFLRLTGYYRKFIKDYGKLAPRPLTDLTKND-GFKW 18
>NP334778 reverse transcriptase [Glycine max]
Length = 431
Score = 102 bits (253), Expect = 2e-21
Identities = 52/143 (36%), Positives = 86/143 (59%)
Frame = +3
Query: 560 RLCVDYRQLNKVTIKNRYPLPRIDDLMDQLKGARVFSKIDLRSGYHQIRVKSDDVQKTAF 619
R+CVDYR LN+ + K+ +PLP ID LM + +FS +D SGY+QI++ +D++KT F
Sbjct: 3 RMCVDYRDLNRASPKDNFPLPHIDILMANMASFALFSFMDGFSGYNQIKMAPEDMEKTTF 182
Query: 620 RTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYLDKFVIVFIDDILIYSKSKEEHVEHM 679
T +G + Y VM FG+ N A + M +F + K + ++D+++ S+ +EEH+ ++
Sbjct: 183 ITLWGTFCYKVMSFGLKNFGATYHRAMVALFQDMMHKEIEAYVDEMIAKSRMEEEHLVNL 362
Query: 680 QVVLKVLKDRKLYAKLSKCEFWL 702
Q + L+ +L KC F L
Sbjct: 363 QNLFGQLRKYRLRLNPRKCVFGL 431
>BQ273711
Length = 409
Score = 95.9 bits (237), Expect = 1e-19
Identities = 42/99 (42%), Positives = 60/99 (60%)
Frame = -2
Query: 17 EQQNQPAEDDVYKGIDKFLKRNPPLFDGGYDPEGANRWLRKIEQIYESLPTSEDRMIAYA 76
E+ +PAE Y+G+ F K +PP F G YDPEGA WL + E+I+E++ E+ + YA
Sbjct: 288 ERDVEPAE---YRGLMAFRKNHPPKFSGDYDPEGARLWLAETEKIFEAMGCLEEHKVLYA 118
Query: 77 SYLFHEEARNWWVHAKSRITPPDGVLTWSIFKEAFLEKY 115
+++ EA NWW K P GV+ W+ FK+ FLE Y
Sbjct: 117 TFMLQGEAENWWKFVKPSFVAPGGVIPWNAFKDKFLENY 1
>TC233837 similar to UP|Q6WAY3 (Q6WAY3) Gag/pol polyprotein, partial (6%)
Length = 402
Score = 93.6 bits (231), Expect = 6e-19
Identities = 48/133 (36%), Positives = 82/133 (61%)
Frame = +2
Query: 595 FSKIDLRSGYHQIRVKSDDVQKTAFRTRYGHYEYLVMPFGVTNAPAIFMDYMNRIFHPYL 654
FS +D SGY+QI + +DV+KT F T +G + Y VM FG+ N A + M +FH +
Sbjct: 2 FSFMDGFSGYNQI*MAREDVEKTTFVTLWGTFSYRVMAFGLKNTGATYQRAMVALFHDMM 181
Query: 655 DKFVIVFIDDILIYSKSKEEHVEHMQVVLKVLKDRKLYAKLSKCEFWLEQVQFLGHVVSE 714
K + V++DD++ S+++ EH+ ++ + L+ +L +KC F ++ + LG +VS+
Sbjct: 182 HKEIEVYVDDMIAKSRTETEHLVNLCKLFGRLQKYQLKLNPTKCTFGVKSGKLLGFIVSQ 361
Query: 715 DGIAVDPAKVEAV 727
GI +DP KV+A+
Sbjct: 362 KGIEIDPEKVKAL 400
>BQ299538
Length = 426
Score = 58.9 bits (141), Expect(2) = 4e-18
Identities = 35/94 (37%), Positives = 50/94 (52%)
Frame = +3
Query: 1172 GTRLSLSSAYHPQSDGQSERTIQTLEDMLRACVLDYKGSWEDFLPLAEFSYNNSYHSSLG 1231
GT L +S++YHP DGQ+ LE LR V D +L AE+ YN ++H+S G
Sbjct: 144 GTYLKMSTSYHP*IDGQTVN--HCLETFLRCFVADQPKM*VQWLSWAEYWYNTNFHASTG 317
Query: 1232 MAPFEALYGRRCKTPLCWLSGEDKITLGPELLQE 1265
PFE +YGR+ +L GE ++ LQ+
Sbjct: 318 TTPFEVVYGRKPPVLNRFLPGEVRVEAVRRELQD 419
Score = 52.4 bits (124), Expect(2) = 4e-18
Identities = 22/40 (55%), Positives = 29/40 (72%)
Frame = +1
Query: 1128 FPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSL 1167
+ + LA I+ KE+V LHGVPA+++SD DP FVS FW L
Sbjct: 13 YSARVLAEIFTKEVVHLHGVPASVLSDEDPIFVSSFWKEL 132
>BQ627806
Length = 435
Score = 58.2 bits (139), Expect(2) = 7e-16
Identities = 29/68 (42%), Positives = 40/68 (58%)
Frame = +1
Query: 843 THDMELAAVVFALKIWRHYLYGVKFTIYSDHQSLKYLFDQKTLNMRQRRWVEFLEDYDFK 902
T+ ELAA+ A+K WR YL G F I +DH+SLK L Q Q+ ++ L +D+
Sbjct: 232 TYVRELAAITVAVKKWRQYLLGHHFVILTDHRSLKELMSQAVQTPEQQIYLARLMGFDYT 411
Query: 903 LQYHPGKA 910
+QY GKA
Sbjct: 412 IQYRAGKA 435
Score = 45.4 bits (106), Expect(2) = 7e-16
Identities = 23/57 (40%), Positives = 33/57 (57%)
Frame = +3
Query: 773 FVWTEKCE*SFQTLKERLTKAPVLTLPDPSKDYDVYCDASKSGLGCVLMQERKVIAY 829
F W E+ + +F LK L +APVL LPD + + V DAS G+G +L Q +A+
Sbjct: 21 FHWNEEADRAFSQLKLALCQAPVLGLPDFNSSFVVETDASGIGMGAILSQNHHPLAF 191
>BI317638 weakly similar to GP|9294238|dbj| contains similarity to reverse
transcriptase~gene_id:K11J14.5 {Arabidopsis thaliana},
partial (5%)
Length = 420
Score = 78.2 bits (191), Expect(2) = 4e-15
Identities = 40/109 (36%), Positives = 62/109 (56%)
Frame = -2
Query: 1059 SCLTCQKVKGEHQKPSGSLQPLSIPEWKWEGISMDFVSGLPRTTTGHDAIWVIVDRLTKS 1118
+CL CQ K E ++ L PL +P WE +S+DF++GL H AI V+VD +K
Sbjct: 329 NCLDCQHTKYETKRIVDLLCPLLVPHRPWEDLSLDFITGLLPYHV-HTAILVVVDHFSKG 153
Query: 1119 AHFIAVNMTFPSEKLARIYVKEIVRLHGVPANIVSDRDPRFVSKFWGSL 1167
H + + + +A +++ + +LHG+P ++VSD D FVS FW L
Sbjct: 152 IHLGMLPSSHTAHTVACLFIDSVAKLHGLPRSLVSDCDLLFVSHFWQEL 6
Score = 22.7 bits (47), Expect(2) = 4e-15
Identities = 9/24 (37%), Positives = 12/24 (49%)
Frame = -1
Query: 1030 HPGTTKMYQDLKKMFWWPGMKKDI 1053
H G K L K +W GM+ D+
Sbjct: 405 HTGIAKTLA*LSKNIYWFGMRTDV 334
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,629,861
Number of Sequences: 63676
Number of extensions: 895814
Number of successful extensions: 4501
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 4405
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4477
length of query: 1435
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1326
effective length of database: 5,698,948
effective search space: 7556805048
effective search space used: 7556805048
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0039.4


