

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.4
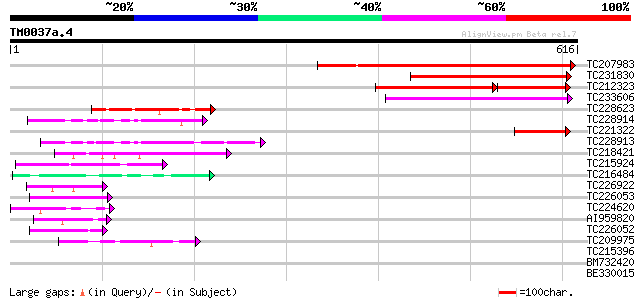
(616 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC207983 448 e-126
TC231830 weakly similar to UP|Q8H2N8 (Q8H2N8) Proteophosphoglyca... 247 1e-65
TC212323 129 3e-44
TC233606 135 5e-32
TC228623 similar to UP|Q9SA21 (Q9SA21) F3O9.2 protein, partial (... 102 7e-22
TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (... 50 2e-06
TC221322 similar to UP|Q9FMG7 (Q9FMG7) Emb|CAB52444.1, partial (7%) 49 7e-06
TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partia... 49 9e-06
TC218421 similar to GB|AAQ65192.1|34365761|BT010569 At3g08670 {A... 46 4e-05
TC215924 similar to GB|AAS68109.1|45592910|BT011785 At2g38360 {A... 45 7e-05
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 45 9e-05
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 45 9e-05
TC226053 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_... 44 2e-04
TC224620 similar to UP|O23254 (O23254) Serine hydroxymethyltrans... 43 5e-04
AI959820 similar to GP|14335118|gb| At1g30200/F12P21_1 {Arabidop... 42 6e-04
TC226052 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_... 42 8e-04
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 42 8e-04
TC215396 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kD chloroplast ... 42 0.001
BM732420 weakly similar to GP|13249030|gb F-box containing prote... 41 0.002
BE330015 41 0.002
>TC207983
Length = 1179
Score = 448 bits (1153), Expect = e-126
Identities = 233/280 (83%), Positives = 257/280 (91%)
Frame = +1
Query: 335 SQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSND 394
SQSL APGSRLPSPSR SV S+SSSRGVSPSRSRPSTPPSRG VSPSRIRPT+SSIQSN+
Sbjct: 1 SQSLFAPGSRLPSPSRTSVLSSSSSRGVSPSRSRPSTPPSRG-VSPSRIRPTNSSIQSNN 177
Query: 395 SVSVLSFIADFRKGKKGAAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLY 454
S+SVL FIADF+KGKKGAA IEDAH+LRLL+NR++QWRFANARAEAVLYIQNAIV+KTLY
Sbjct: 178 SISVLRFIADFKKGKKGAANIEDAHKLRLLHNRYLQWRFANARAEAVLYIQNAIVEKTLY 357
Query: 455 NVWIATLSLWESVTRKKINLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVED 514
NVWI TLSLWESV RK+INLQQLKLELKLNSV+NDQM YLD+WA LE DH+DA+S AVED
Sbjct: 358 NVWITTLSLWESVIRKRINLQQLKLELKLNSVMNDQMTYLDDWAVLERDHIDAVSKAVED 537
Query: 515 LEASTLRLPVTRGAMVDIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVA 574
LEASTLRLPVT GAM DIEHLKVAICQAVDVMQAM SAI SL SQVEGMN+LIS VA +A
Sbjct: 538 LEASTLRLPVTGGAMSDIEHLKVAICQAVDVMQAMASAICSLLSQVEGMNNLISEVAAIA 717
Query: 575 TKEKSILDECEMLLASVAALQVEESSLQTHLIQFKQVLGI 614
+EK++LDECEMLLASVAA+QVEESSL+THL+Q Q LG+
Sbjct: 718 VQEKTMLDECEMLLASVAAMQVEESSLRTHLMQIMQALGM 837
Score = 30.8 bits (68), Expect = 1.9
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 3/53 (5%)
Frame = +1
Query: 65 RCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPS---PPSRRLSTPVNDS 114
R PSP+ T + ++S R +S R RPSTPPS PSR P N S
Sbjct: 28 RLPSPSRTSVLSSSSS------RGVSPSRSRPSTPPSRGVSPSR--IRPTNSS 162
>TC231830 weakly similar to UP|Q8H2N8 (Q8H2N8) Proteophosphoglycan-like,
partial (12%)
Length = 677
Score = 247 bits (631), Expect = 1e-65
Identities = 126/175 (72%), Positives = 148/175 (84%)
Frame = +2
Query: 436 ARAEAVLYIQNAIVQKTLYNVWIATLSLWESVTRKKINLQQLKLELKLNSVLNDQMAYLD 495
A+AE V YIQN +K+LYNVW TLS+WES+ RK+INLQQL+LELKLNS+LNDQMAYLD
Sbjct: 5 AQAEDVFYIQNVTAEKSLYNVWHTTLSIWESIIRKRINLQQLQLELKLNSILNDQMAYLD 184
Query: 496 EWATLETDHVDALSGAVEDLEASTLRLPVTRGAMVDIEHLKVAICQAVDVMQAMGSAIRS 555
+WA LE+DH+D+LSGAVEDLEASTLRLP+T GA DIEHLK AI AVD MQAMGSAI
Sbjct: 185 DWAVLESDHIDSLSGAVEDLEASTLRLPLTGGAKADIEHLKHAIYSAVDGMQAMGSAICP 364
Query: 556 LFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQVEESSLQTHLIQFKQ 610
L SQVEGMN+LIS VAVV+++EK++LDECE LL A+QVEE SL+THL+Q KQ
Sbjct: 365 LLSQVEGMNNLISEVAVVSSREKAMLDECEALLNFATAMQVEEYSLRTHLMQIKQ 529
>TC212323
Length = 765
Score = 129 bits (325), Expect(2) = 3e-44
Identities = 68/133 (51%), Positives = 86/133 (64%)
Frame = +2
Query: 398 VLSFIADFRKGKKGAAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVW 457
VLSF D +GK I DAH LRL +NR +QWRF NARA+A L Q +K+LY+ W
Sbjct: 2 VLSFAVDVSRGKVAENRIFDAHLLRLFHNRLLQWRFVNARADAALSAQTLNAEKSLYDAW 181
Query: 458 IATLSLWESVTRKKINLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEA 517
+A +L ESV K+ Q LK + KL S+L DQM L++WATL+ + +LSGA E L A
Sbjct: 182 VAMSNLRESVRAKRAEFQLLKQQFKLISILKDQMVCLEDWATLDPVYSSSLSGATEALRA 361
Query: 518 STLRLPVTRGAMV 530
STLRLPV GA +
Sbjct: 362 STLRLPVVGGAKI 400
Score = 67.8 bits (164), Expect(2) = 3e-44
Identities = 36/79 (45%), Positives = 53/79 (66%)
Frame = +1
Query: 531 DIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLAS 590
D+ +LK AI A+DVMQAM S+I L +V +N L+ VA + KE+ +L+EC+ LL+
Sbjct: 397 DLVNLKDAISSAMDVMQAMASSICLLSPKVGQLNSLVVEVANLTAKERVLLEECKDLLSV 576
Query: 591 VAALQVEESSLQTHLIQFK 609
+ +QV E SL+TH+ Q K
Sbjct: 577 ITTMQVRECSLRTHVAQLK 633
>TC233606
Length = 871
Score = 135 bits (340), Expect = 5e-32
Identities = 76/203 (37%), Positives = 122/203 (59%)
Frame = +1
Query: 409 KKGAAFIEDAHQLRLLYNRFMQWRFANARAEAVLYIQNAIVQKTLYNVWIATLSLWESVT 468
+KG++ ED H LRLLYNR++QWRFANA+A +V+ Q QK LY+ + + +SV
Sbjct: 7 EKGSSHQEDVHSLRLLYNRYLQWRFANAKAHSVMKAQQTESQKALYSQAMRISEMRDSVN 186
Query: 469 RKKINLQQLKLELKLNSVLNDQMAYLDEWATLETDHVDALSGAVEDLEASTLRLPVTRGA 528
+K+I L+ L+ L+++L Q+ YLDEW+T+ ++ +++ A++ L ++ RLPV
Sbjct: 187 KKRIELELLRRSKTLSTILEAQIPYLDEWSTMMEEYSVSITEAIQALVNASERLPVGGNV 366
Query: 529 MVDIEHLKVAICQAVDVMQAMGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLL 588
VD+ L A+ A +M+ M S I+ + E + IS +A VA E++++ EC LL
Sbjct: 367 RVDVRQLGEALNSASKMMETMISNIQRFMPKAEETDVSISELARVAGGERALVGECGDLL 546
Query: 589 ASVAALQVEESSLQTHLIQFKQV 611
+ Q+EE SL+ LIQ V
Sbjct: 547 SKTYKSQLEECSLRGQLIQLHSV 615
>TC228623 similar to UP|Q9SA21 (Q9SA21) F3O9.2 protein, partial (85%)
Length = 1082
Score = 102 bits (253), Expect = 7e-22
Identities = 65/138 (47%), Positives = 83/138 (60%), Gaps = 4/138 (2%)
Frame = +3
Query: 90 SAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQ 149
SAERKRP+TPPSPPS STPV DS++D LSSR+++ SR+PE LWPS MRSLSVSFQ
Sbjct: 9 SAERKRPATPPSPPSP--STPVQDSSVDVHLSSRRLSGSRMPEA--LWPSRMRSLSVSFQ 176
Query: 150 SDIISIPVSKKE----RPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKN 205
SD ISIPV KKE R + R + A ++ E + RKA R L +
Sbjct: 177 SDTISIPVIKKEDQYWREAEGSKSRAAKKKEEEAEKRAE--AAARKAEARR----LAEQE 338
Query: 206 ASDQSENSKPVDSLPSRL 223
+ ++ K VD +R+
Sbjct: 339 EKELEKSMKKVDKKATRV 392
>TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (Dach1),
partial (3%)
Length = 784
Score = 50.4 bits (119), Expect = 2e-06
Identities = 57/202 (28%), Positives = 83/202 (40%), Gaps = 6/202 (2%)
Frame = +3
Query: 20 PRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAA 79
PR P + T TAT R + + + +S +PPS+P PSP+ T T++
Sbjct: 48 PRAPPPHRSQTPTRTTTATRRTTTSTLPPASPRSSSPPSSP------PSPSTTPTSSAPG 209
Query: 80 SLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPS 139
+ P S SA ++RP TP P S ++P S +S + A S+ P WPS
Sbjct: 210 N---APHSSPSASKRRP-TPSGPSSAASTSPKPTS------TSSRAAPSKSP---FTWPS 350
Query: 140 TMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQGET-----PSTIRKATP 194
SL S S P + ++ S + AS+ A+ T PS + TP
Sbjct: 351 A--SLGTSMSSPASPPPPAPNASTSSTTSAASPALASSAANTASATTAPSLPSIPSRTTP 524
Query: 195 ERKRS-PLKGKNASDQSENSKP 215
RS P S S + P
Sbjct: 525 TTARSTPSSSNPTSSTSPTATP 590
>TC221322 similar to UP|Q9FMG7 (Q9FMG7) Emb|CAB52444.1, partial (7%)
Length = 665
Score = 48.9 bits (115), Expect = 7e-06
Identities = 24/61 (39%), Positives = 39/61 (63%)
Frame = +2
Query: 549 MGSAIRSLFSQVEGMNDLISGVAVVATKEKSILDECEMLLASVAALQVEESSLQTHLIQF 608
M S+I L +V +N L+ V ++ KE+ +L+EC LL+++ +QV E SL+TH+ Q
Sbjct: 2 MASSICLLSPKVGQLNSLVVXVXNLSAKERVLLEECRDLLSAITTMQVRECSLRTHVAQL 181
Query: 609 K 609
K
Sbjct: 182 K 184
>TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (19%)
Length = 692
Score = 48.5 bits (114), Expect = 9e-06
Identities = 69/246 (28%), Positives = 100/246 (40%), Gaps = 1/246 (0%)
Frame = +2
Query: 34 TATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAER 93
T TAT R + + + +S +PPS+P PSP+ T T++ A+ P S SA
Sbjct: 29 TTTATRRTTTSTLPPASPRSSSPPSSP------PSPSTTPTSSAPAN---APPSSPSAS- 178
Query: 94 KRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDII 153
R TP P S ++P S +S + A S+ P + WPS SL+ S S
Sbjct: 179 TRLQTPSGPSSDASTSPKPTS------TSSRAAPSKSP---STWPSA--SLATSMSSPAS 325
Query: 154 SIPVSKKERPVTSASDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNASDQSENS 213
P ++ S + AS+VA+ T + +TP +P + S
Sbjct: 326 PQPRVPSASTSSTTSAASPASASSVANTASVTTAPSPPSTPSTTTTP-------PPTARS 484
Query: 214 KPVDSLPSRLIDQHRWPSRIGGKVSSSALNRSVDFGDAR-MLKTPASGTGFSSLRRLSLS 272
P S P+ P R +VSS + S +R K P + S R
Sbjct: 485 TPSSSNPTLSTSPTATPKRT--RVSSPTPSSSSTSRSSRPSPKEPTATVTVSHTR----G 646
Query: 273 EEASKP 278
EEASKP
Sbjct: 647 EEASKP 664
Score = 38.9 bits (89), Expect = 0.007
Identities = 28/95 (29%), Positives = 44/95 (45%), Gaps = 3/95 (3%)
Frame = +2
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPA 78
+P+P + A ++ +A + A S S +P+PPSTPS + +P T +TP+
Sbjct: 323 SPQPRVPSASTSSTTSAASPASASSVANTASVTTAPSPPSTPSTT---TTPPPTARSTPS 493
Query: 79 AS---LQLLPKRSLSAERKRPSTPPSPPSRRLSTP 110
+S L P + R TP S + R S P
Sbjct: 494 SSNPTLSTSPTATPKRTRVSSPTPSSSSTSRSSRP 598
>TC218421 similar to GB|AAQ65192.1|34365761|BT010569 At3g08670 {Arabidopsis
thaliana;} , partial (46%)
Length = 1256
Score = 46.2 bits (108), Expect = 4e-05
Identities = 60/210 (28%), Positives = 86/210 (40%), Gaps = 17/210 (8%)
Frame = +1
Query: 49 SRYKSPTPPSTPSGSRRCPS-----PNLTRTATPAASLQLLPKRSLSAERKRPSTP---P 100
S Y P+ P T S S PS P +R++TP+ + S +AER RPS+ P
Sbjct: 394 SSYIRPSSPITRSSSSARPSTPSSRPTASRSSTPSKPRPV--STSSTAERNRPSSQGSRP 567
Query: 101 SPPSRRLSTPVN---DSAIDGRLSSRKVAASRLPEGHNLWPS---TMRSLSVSFQSDIIS 154
S PS R P N SA R SR +R +L PS T+ SL+ + +
Sbjct: 568 STPSSRPHIPANLHSPSASSTRSLSRPSTPTRRSSMPSLSPSPSPTIGSLTSAGRVSSNG 747
Query: 155 IPVSKKERPVTSASDRTLRPASNVAHRQG--ETPSTIRKATPERKRSPLKGK-NASDQSE 211
+ RP +S S R P + ETP +R P+R S + +
Sbjct: 748 RNSAPASRP-SSPSPRVRPPPQPIVPPDFPLETPPNLRTTLPDRPVSAGRSRPGGVTMKT 924
Query: 212 NSKPVDSLPSRLIDQHRWPSRIGGKVSSSA 241
NS + P + +H P G+V+ A
Sbjct: 925 NSSETQASPVTMPRRHSSPIVSRGRVTEPA 1014
Score = 41.2 bits (95), Expect = 0.001
Identities = 43/145 (29%), Positives = 65/145 (44%), Gaps = 13/145 (8%)
Frame = +1
Query: 261 TGFSSLRRLSLSEE-----ASKPLQRAS------SDSVRLLSLVASGRTGSEVKSVDDCS 309
T S RL++S+ AS+P + +S S S S +S R S + + S
Sbjct: 211 TSTSKTSRLAVSQSENNNPASRPARSSSVTRSSISTSHSQYSSYSSNRHSSSILNTSSAS 390
Query: 310 LHD-LRPHRSSTPHTDRAGLSIAGVRSQSLSAPGSRLPS-PSRISVQSTSSSRGVSPSRS 367
+ +RP S+P T + + S +A S PS P +S ST+ S S
Sbjct: 391 VSSYIRP---SSPITRSSSSARPSTPSSRPTASRSSTPSKPRPVSTSSTAERNRPSSQGS 561
Query: 368 RPSTPPSRGGVSPSRIRPTSSSIQS 392
RPSTP SR + + P++SS +S
Sbjct: 562 RPSTPSSRPHIPANLHSPSASSTRS 636
Score = 37.4 bits (85), Expect = 0.020
Identities = 63/237 (26%), Positives = 87/237 (36%), Gaps = 16/237 (6%)
Frame = +1
Query: 174 PASNVAHRQGETPSTIRKATPERKRSPLKGKN---ASDQSENSKPVDSLPSRLIDQHRWP 230
P + V +GE+ +T+ RS K A QSEN+ P S P+R R
Sbjct: 133 PGTPVFPSEGESQTTLAPPRRSLTRSTSTSKTSRLAVSQSENNNPA-SRPARSSSVTRSS 309
Query: 231 -SRIGGKVSSSALNRSVDFGDARMLKTPASGTGF-----SSLRRLSLSEEASKPLQRASS 284
S + SS + NR + +L T ++ S + R S S S P R ++
Sbjct: 310 ISTSHSQYSSYSSNRH----SSSILNTSSASVSSYIRPSSPITRSSSSARPSTPSSRPTA 477
Query: 285 DSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRSSTPHTDRAGLSIAGVRSQSLSAPG-- 342
S T S + S SS PH S + ++SLS P
Sbjct: 478 SRSSTPSKPRPVSTSSTAERNRPSSQGSRPSTPSSRPHIPANLHSPSASSTRSLSRPSTP 657
Query: 343 ---SRLPS--PSRISVQSTSSSRGVSPSRSRPSTPPSRGGVSPSRIRPTSSSIQSND 394
S +PS PS + +S G S R S P SR R+RP I D
Sbjct: 658 TRRSSMPSLSPSPSPTIGSLTSAGRVSSNGRNSAPASRPSSPSPRVRPPPQPIVPPD 828
>TC215924 similar to GB|AAS68109.1|45592910|BT011785 At2g38360 {Arabidopsis
thaliana;} , partial (68%)
Length = 947
Score = 45.4 bits (106), Expect = 7e-05
Identities = 47/169 (27%), Positives = 71/169 (41%), Gaps = 4/169 (2%)
Frame = +3
Query: 7 QQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTP-PSTPSGSRR 65
Q A+ RL+ P P + TA+ +A R R+ R +SP+P P PS S R
Sbjct: 108 QAAAPSRLRRTTRPSAPSSTTSPRPSDTASTSAARGRSSGTAPRSRSPSPSPKPPSASAR 287
Query: 66 CPSPNLTRTATPA--ASLQLLPKRSLS-AERKRPSTPPSPPSRRLSTPVNDSAIDGRLSS 122
SP T TP+ + Q L LS + S+PP P S +N S L
Sbjct: 288 -TSPTSASTTTPSFHSFSQSLSSPILSLSSSSSASSPPGPSSTSSVRQINHSLFLAEL-- 458
Query: 123 RKVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRT 171
++ + P + S S S D+ S P+S P ++++ R+
Sbjct: 459 -----FQISKP*RSSPPSPSSSSSSPALDLFSSPLSCLASPSSASTARS 590
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 45.1 bits (105), Expect = 9e-05
Identities = 51/219 (23%), Positives = 72/219 (32%)
Frame = +1
Query: 4 CESQQASSRRLKAVETPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGS 63
C S SRR + P P T + + +S S SP+P S P +
Sbjct: 49 CSSSGHCSRRRCSSSPPSP---------TRTTSPASSQSTPSSPPSTTTSPSPTSPPKST 201
Query: 64 RRCPSPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSR 123
+ PSP+ T P + STPPS PSR S + S SS
Sbjct: 202 AKPPSPSAPSTTPPCRTFSR-------------STPPSTPSRTSSPSTSSSTTSAPRSS- 339
Query: 124 KVAASRLPEGHNLWPSTMRSLSVSFQSDIISIPVSKKERPVTSASDRTLRPASNVAHRQG 183
+R P P + P + P +S S ++ A +
Sbjct: 340 ----TRSPTAPPSPPQCTKP------------PAPPPDPPASSTS------PTSTAEKSH 453
Query: 184 ETPSTIRKATPERKRSPLKGKNASDQSENSKPVDSLPSR 222
P T +P+R +P K + Q S LP R
Sbjct: 454 SPPKTTTAHSPQRSSNP*KKSLTTSQ*SRSAKFSPLPQR 570
Score = 31.2 bits (69), Expect = 1.4
Identities = 36/118 (30%), Positives = 47/118 (39%), Gaps = 2/118 (1%)
Frame = +1
Query: 259 SGTGFSSLRRLSLSEEASKPLQRASSDSVRLLSLVASGRTGSEVKSVDDCSLHDLRPHRS 318
S +G S RR S S + R S +S T S S P +
Sbjct: 52 SSSGHCSRRRCSSSPPSP----------TRTTSPASSQSTPSSPPSTTTSPSPTSPPKST 201
Query: 319 STPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSP--SRSRPSTPPS 374
+ P + A + R+ S S P S +PSR S STSSS +P S P+ PPS
Sbjct: 202 AKPPSPSAPSTTPPCRTFSRSTPPS---TPSRTSSPSTSSSTTSAPRSSTRSPTAPPS 366
Score = 28.9 bits (63), Expect = 7.0
Identities = 21/70 (30%), Positives = 31/70 (44%)
Frame = +1
Query: 319 STPHTDRAGLSIAGVRSQSLSAPGSRLPSPSRISVQSTSSSRGVSPSRSRPSTPPSRGGV 378
S PH S R + S+P S + S S QST SS + + P++PP
Sbjct: 28 SLPHLP*CSSSGHCSRRRCSSSPPSPTRTTSPASSQSTPSSPPSTTTSPSPTSPPKSTAK 207
Query: 379 SPSRIRPTSS 388
PS P+++
Sbjct: 208 PPSPSAPSTT 237
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 45.1 bits (105), Expect = 9e-05
Identities = 34/102 (33%), Positives = 46/102 (44%), Gaps = 14/102 (13%)
Frame = +2
Query: 19 TPRPPLVLAEKNNAATATAT-ARRSRTR------EVTSRYKSPTPPSTPSGSRRCPS--- 68
TP PP + AT TA + +S TR S +PTPP + SR+ PS
Sbjct: 197 TPLPPPTSRSSPSLATETAAPSTKSATRPPPPPTRSRSSTPTPTPPGAAAPSRKPPSSAA 376
Query: 69 ----PNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRR 106
P L+ + P+ S + + S S PS PPSPP+ R
Sbjct: 377 LPTAPTLSASTAPSKSPPVTWRSSWSTWTAAPSRPPSPPAAR 502
Score = 37.7 bits (86), Expect = 0.015
Identities = 41/173 (23%), Positives = 67/173 (38%), Gaps = 4/173 (2%)
Frame = +2
Query: 20 PRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAA 79
P+PP + A + + RS ++ PP P+ P P T ++P+
Sbjct: 59 PQPP*WPSSATAATPTSVSPSRSPPSAASASPSPSLPPPPPNQPPATPLPPPTSRSSPSL 238
Query: 80 SLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDSAIDGRLSSRKVAASRLPEGHNLWPS 139
+ + + SA R PP P R STP S + +++ LP L S
Sbjct: 239 ATETAAPSTKSATRP----PPPPTRSRSSTPTPTPPGAAAPSRKPPSSAALPTAPTLSAS 406
Query: 140 TMRSLS--VSFQS--DIISIPVSKKERPVTSASDRTLRPASNVAHRQGETPST 188
T S S V+++S + S+ P + S ++ P V +G ST
Sbjct: 407 TAPSKSPPVTWRSSWSTWTAAPSRPPSPPAARSRKSDSPRWRVTSSRGSRTST 565
Score = 28.9 bits (63), Expect = 7.0
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 13/70 (18%)
Frame = +2
Query: 341 PGSRLPSPSRISVQS--TSSSRGVSPSRSRPSTPPSR-----------GGVSPSRIRPTS 387
P + LP P+ S S T ++ + S +RP PP+R G +PSR P+S
Sbjct: 191 PATPLPPPTSRSSPSLATETAAPSTKSATRPPPPPTRSRSSTPTPTPPGAAAPSRKPPSS 370
Query: 388 SSIQSNDSVS 397
+++ + ++S
Sbjct: 371 AALPTAPTLS 400
>TC226053 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_13), partial
(26%)
Length = 1335
Score = 44.3 bits (103), Expect = 2e-04
Identities = 31/94 (32%), Positives = 44/94 (45%), Gaps = 4/94 (4%)
Frame = +2
Query: 22 PPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSG--SRRCPSPNLTRTATPAA 79
PP ++ + A T++A + ++ SP PP T SG S R +P + T+ P
Sbjct: 332 PPPTISLPSATAPRTSSASP*PSSSASAAAPSPPPPCTSSGPSSPRATAPPPSTTSPPTT 511
Query: 80 SLQLLPKRSLSAERKRP--STPPSPPSRRLSTPV 111
+ RSL+ R RP PPSPP RL V
Sbjct: 512 KTRSRAPRSLATSRFRPLRPLPPSPPQLRLQPRV 613
>TC224620 similar to UP|O23254 (O23254) Serine hydroxymethyltransferase
(Serine methylase) (Glycine hydroxymethyltransferase)
(SHMT) , complete
Length = 1887
Score = 42.7 bits (99), Expect = 5e-04
Identities = 37/119 (31%), Positives = 51/119 (42%), Gaps = 5/119 (4%)
Frame = +1
Query: 1 MDVCESQQASSRRLKAVETPRPPLVLAEKNNA----ATATATARRSRTREVTSRYKSPTP 56
++ ES + R + RP L+ N ATAT A + TR TS +P+
Sbjct: 169 VNAAESSSSPPRTSPPSPSSRPSAALSRTNTPRACRATATTAAMNTSTRSKTSAAHAPSK 348
Query: 57 PSTPSGSRRCPSPNL-TRTATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVNDS 114
PST + P+P T + TPA RP++PP+PPS STP S
Sbjct: 349 PSTST-----PNPGASTSSPTPA---------------PRPTSPPTPPS---STPTTAS 456
>AI959820 similar to GP|14335118|gb| At1g30200/F12P21_1 {Arabidopsis
thaliana}, partial (19%)
Length = 342
Score = 42.4 bits (98), Expect = 6e-04
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 4/88 (4%)
Frame = +2
Query: 27 AEKNNAATATATARRSRTREVTSRYKSPT----PPSTPSGSRRCPSPNLTRTATPAASLQ 82
A ++ A +T ++RRS+T S SPT PP P S PS + +++ +
Sbjct: 26 AAASSRAASTPSSRRSKTSSSASTASSPTTTPLPPPPPPTSPAAPSGTSSASSSAES*NP 205
Query: 83 LLPKRSLSAERKRPSTPPSPPSRRLSTP 110
P S SA P+ PP PPSR L P
Sbjct: 206 SKPSVSSSA----PNAPPPPPSRPLPPP 277
>TC226052 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (At1g65720/F1E22_13), partial
(39%)
Length = 941
Score = 42.0 bits (97), Expect = 8e-04
Identities = 31/89 (34%), Positives = 43/89 (47%), Gaps = 4/89 (4%)
Frame = +1
Query: 22 PPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSG--SRRCPSPNLTRTATPAA 79
PP ++ + A T++A S + V++ SP PP T SG S R P + T+ P
Sbjct: 391 PPRTISPPSATAPRTSSASPSPSSSVSAAAPSPPPPCTSSGPSSPRATPPPPSTTSPPTK 570
Query: 80 SLQLLPKRSLSAERKRPSTPP--SPPSRR 106
+ P RS + R P PP SPP RR
Sbjct: 571 TRSRAP-RS*ATSRFLPPRPPLLSPPLRR 654
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 42.0 bits (97), Expect = 8e-04
Identities = 44/160 (27%), Positives = 68/160 (42%), Gaps = 6/160 (3%)
Frame = +1
Query: 54 PTPPSTPSGSRRCP-SPNLTRTATPAASLQLLPKRSLSAERKRPSTPPSPPSR--RLSTP 110
PTPP TPS S + P +P +T +P + +P TPPSP S+ ++TP
Sbjct: 121 PTPPKTPSPSSQPPFTPTPPKTPSPTSQPPYIP------------TPPSPISQPPSIATP 264
Query: 111 VNDSAIDGRLSSRKVAASRLPEGHNLWPSTMRSLSVSFQSDI---ISIPVSKKERPVTSA 167
N + +S+ S +P +L PS S S S P + P TSA
Sbjct: 265 PNTLS----PTSQPPYPSTIPPSTSLSPSYPPSPSPSSAPTYPPSYLAPATSPPSPSTSA 432
Query: 168 SDRTLRPASNVAHRQGETPSTIRKATPERKRSPLKGKNAS 207
+ T+ PA+ +PS+ + + +P + AS
Sbjct: 433 TTPTISPAATT-----PSPSSSSSSGASNETTPARPNGAS 537
>TC215396 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kD chloroplast nucleoid DNA
binding protein (CND41), partial (69%)
Length = 1808
Score = 41.6 bits (96), Expect = 0.001
Identities = 30/106 (28%), Positives = 47/106 (44%), Gaps = 11/106 (10%)
Frame = +1
Query: 16 AVETPRPPLVLAEKNNAATATATARRSRTREV---TSRYKSPTPPSTPSGSRRCPSPNLT 72
A +T R LV ++A+ AT + ++ +S SP PP+ P+ S P P T
Sbjct: 796 AAKTTRASLVAPPASSASAATPSPSSNKPPPYIVKSSPTASPPPPAPPAASPSAPPPLPT 975
Query: 73 RT--------ATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTP 110
+ A P ++ P +A + PPSPP+ + STP
Sbjct: 976 SSTPPSPPSPAAPPSTASTSPASPWAAPNSQSPPPPSPPAVQSSTP 1113
Score = 34.3 bits (77), Expect = 0.17
Identities = 25/103 (24%), Positives = 45/103 (43%), Gaps = 9/103 (8%)
Frame = +1
Query: 19 TPRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRT---- 74
+P PP ++ T+++ A ++ + + S +TPS S P P + ++
Sbjct: 751 SP*PP-----PTSSTTSSSVAAKTTRASLVAPPASSASAATPSPSSNKPPPYIVKSSPTA 915
Query: 75 -----ATPAASLQLLPKRSLSAERKRPSTPPSPPSRRLSTPVN 112
A PAAS P S+ P +P +PPS ++P +
Sbjct: 916 SPPPPAPPAASPSAPPPLPTSSTPPSPPSPAAPPSTASTSPAS 1044
Score = 32.7 bits (73), Expect = 0.49
Identities = 35/126 (27%), Positives = 55/126 (42%), Gaps = 7/126 (5%)
Frame = +1
Query: 20 PRPPLVLAEKNNAATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAA 79
P PP A + A+T+ A+ + + SP PPS P+ P+P +PA+
Sbjct: 988 PSPPSPAAPPSTASTSPASPWAAPNSQ------SPPPPSPPAVQSSTPAP*-----SPAS 1134
Query: 80 SLQLLPKRSLSAERKRPSTPP---SPPSRR-LSTPVNDSAIDGRLSSRKVAASRLP---E 132
P + + + P+TPP SP S R + + S + R AASR +
Sbjct: 1135 PPPPTPPSAPPSGKACPNTPPPGNSPYSTRVMISAATRSFLFPR*IFPSPAASRCSFRRK 1314
Query: 133 GHNLWP 138
G++ WP
Sbjct: 1315 GYSTWP 1332
Score = 28.5 bits (62), Expect = 9.2
Identities = 23/65 (35%), Positives = 25/65 (38%), Gaps = 16/65 (24%)
Frame = +1
Query: 54 PTPPSTPSGSRRCP-------SPNLTRTATPAASLQLLPKR---------SLSAERKRPS 97
PTPPS P + CP SP TR AA+ L R S RK S
Sbjct: 1144 PTPPSAPPSGKACPNTPPPGNSPYSTRVMISAATRSFLFPR*IFPSPAASRCSFRRKGYS 1323
Query: 98 TPPSP 102
T P P
Sbjct: 1324 TWPVP 1338
>BM732420 weakly similar to GP|13249030|gb F-box containing protein TIR1
{Populus tremula x Populus tremuloides}, partial (19%)
Length = 425
Score = 40.8 bits (94), Expect = 0.002
Identities = 22/51 (43%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Frame = +1
Query: 55 TPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAERKRPS-TPPSPPS 104
TPPST S S CPSP T ++P S S +A+ P +PPSPP+
Sbjct: 13 TPPSTNSTSNACPSPTTTSPSSPTPSPPSKTSFSRAAKASAPPLSPPSPPT 165
>BE330015
Length = 479
Score = 40.8 bits (94), Expect = 0.002
Identities = 27/80 (33%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Frame = +3
Query: 33 ATATATARRSRTREVTSRYKSPTPPSTPSGSRRCPSPNLTRTATPAASLQLLPKRSLSAE 92
A A A+++ + + Y SP P+ P+G R SP +TATPAA
Sbjct: 27 AAAPASSKTTPATPTSPTYASPPTPTAPAGERN-RSPATAKTATPAA------------- 164
Query: 93 RKRPSTPP--SPPSRRLSTP 110
RPS P +PP+RR ++P
Sbjct: 165 --RPSNPTERTPPARRTTSP 218
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.125 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,766,903
Number of Sequences: 63676
Number of extensions: 389136
Number of successful extensions: 6659
Number of sequences better than 10.0: 786
Number of HSP's better than 10.0 without gapping: 4690
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6112
length of query: 616
length of database: 12,639,632
effective HSP length: 103
effective length of query: 513
effective length of database: 6,081,004
effective search space: 3119555052
effective search space used: 3119555052
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0037a.4


