

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
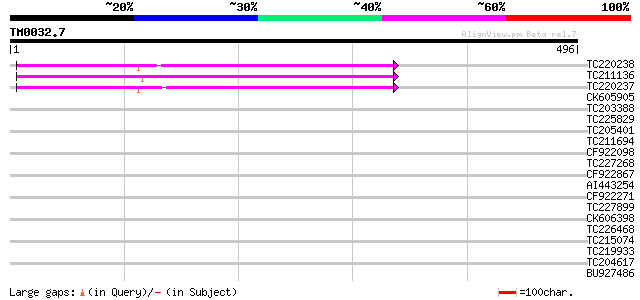
Query= TM0032.7
(496 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220238 UP|Q84VH5 (Q84VH5) Envelope-like protein (Fragment), co... 270 1e-72
TC211136 UP|Q84VI3 (Q84VI3) Envelope-like protein (Fragment), co... 263 2e-70
TC220237 UP|O64406 (O64406) Envelope-like (Fragment), complete 262 2e-70
CK605905 33 0.22
TC203388 homologue to UP|H2B_GOSHI (O22582) Histone H2B, complete 33 0.22
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 33 0.38
TC205401 similar to GB|AAP37850.1|30725656|BT008491 At4g26630 {A... 32 0.50
TC211694 weakly similar to UP|WAIP_HUMAN (O43516) Wiskott-Aldric... 32 0.65
CF922098 32 0.65
TC227268 32 0.85
CF922867 32 0.85
AI443254 weakly similar to GP|9759050|dbj| AAA-type ATPase-like ... 31 1.1
CF922271 31 1.1
TC227899 31 1.1
CK606398 31 1.1
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 31 1.5
TC215074 similar to UP|O04237 (O04237) Transcription factor, par... 31 1.5
TC219933 weakly similar to UP|Q9EQF1 (Q9EQF1) GABA-A epsilon sub... 31 1.5
TC204617 similar to UP|Q9ASZ9 (Q9ASZ9) At1g60730/F8A5_24, partia... 31 1.5
BU927486 weakly similar to GP|20804751|dbj P0698A10.6 {Oryza sat... 30 1.9
>TC220238 UP|Q84VH5 (Q84VH5) Envelope-like protein (Fragment), complete
Length = 2128
Score = 270 bits (689), Expect = 1e-72
Identities = 144/340 (42%), Positives = 200/340 (58%), Gaps = 6/340 (1%)
Frame = +1
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E + P ED T+EE+ E++P P+ PG + + SD+E P
Sbjct: 571 SSTPNAEVLSSPSKEDSTEEEDQATEETPAPRAPEPAPGDLIDLEEVESDEEPIANKLAP 750
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP------VDVEDSESD 120
+ R P+ R T+ K + T S + K +P + DS+ D
Sbjct: 751 GIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPTTSRRSKVAIPSKKRKEISSSDSDDD 930
Query: 121 AEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGP 180
E DV DI+ + K SGK++P NVP P+D +SFH ++V++WKFV + R+A RE+G
Sbjct: 931 VELDVPDIK---RAKTSGKKVPGNVPDAPLDNISFHSIDNVERWKFVYQRRLALERELGR 1101
Query: 181 DVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVEC 240
D L+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +C
Sbjct: 1102DALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVFVRGKC 1281
Query: 241 VVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILN 300
V FSPAVIN+ LGR D VS +AKE+TA V+ WP K LS G LSVKYAIL+
Sbjct: 1282VRFSPAVINKYLGRPTDGVVDITVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAILH 1461
Query: 301 RIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
RI A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 1462RIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 1581
>TC211136 UP|Q84VI3 (Q84VI3) Envelope-like protein (Fragment), complete
Length = 2043
Score = 263 bits (671), Expect = 2e-70
Identities = 142/337 (42%), Positives = 199/337 (58%), Gaps = 3/337 (0%)
Frame = +1
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E P E T+E++ E++P P+ PG + + SD+E P
Sbjct: 547 SSAPNAEALPSPSEEGSTEEDDQAAEETPAPRAPEPAPGDLIDLEEVESDEEPIANRLAP 726
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDV--EDSESDAEAD 124
+ R P+ R T+ K + S + K +P E S SD++ D
Sbjct: 727 GIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPATSRRSKVAIPSKKRKEISSSDSDKD 906
Query: 125 VQ-DIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGPDVL 183
V+ D+ S+K K SGK++P NVP P+D +SFH +V+KWK+V + R+A RE+G D L
Sbjct: 907 VELDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDAL 1086
Query: 184 ECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVECVVF 243
+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +CV F
Sbjct: 1087DCKEIMDLIKAAGLLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKF 1266
Query: 244 SPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILNRID 303
SPAVIN+ LGR D +VS +AKE+TA V+ WP K LS G LSVKYAIL+RI
Sbjct: 1267SPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIG 1446
Query: 304 AVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
A NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 1447AANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 1557
>TC220237 UP|O64406 (O64406) Envelope-like (Fragment), complete
Length = 1974
Score = 262 bits (670), Expect = 2e-70
Identities = 143/340 (42%), Positives = 197/340 (57%), Gaps = 6/340 (1%)
Frame = +1
Query: 7 SKVQNKEPTNVPHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTP 66
S N E + E+ T+EEE E++P P+ PG + + SD+E P
Sbjct: 508 SSTPNAEVLSSSSKEESTEEEEQATEETPAPRAPEPAPGDLIDLEEVESDEEPIANKLAP 687
Query: 67 EVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP------VDVEDSESD 120
+ R P+T R T+ K + T S K +P DS+ D
Sbjct: 688 GIAERLQSRKGKTPITRSGRIKTMAQKKSTPITPTTSRWSKVAIPSKKRKEFSSSDSDDD 867
Query: 121 AEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRVSFHLEESVQKWKFVCKIRMAKVREVGP 180
E DV DI+ ++K SGK++P NVP P+D +SFH +V++WKFV + R+A RE+G
Sbjct: 868 VELDVPDIKRAKK---SGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALERELGR 1038
Query: 181 DVLECKDVIVLIEKAGLMKTIQNVGRCYEKLVREFLVNLSVDVGLPESDEFRKVFVRVEC 240
D L+CK+++ LI+ AGL+KT+ +G CYE LVREF+VN+ D+ +SDE++KVFVR +C
Sbjct: 1039DALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVFVRGKC 1218
Query: 241 VVFSPAVINQALGRSAIEFADKEVSMDDVAKELTAGHVKKWPSKKLLSTGNLSVKYAILN 300
V FSPAVIN+ LGR D VS +AKE+TA V+ WP K LS G LSVKYAIL+
Sbjct: 1219VRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAILH 1398
Query: 301 RIDAVNWVPTQHTSGVSATLAKLIYMIGKSIAFDFGAFVF 340
RI NWVPT HTS V+ L K +Y +G F+FG ++F
Sbjct: 1399RIGTANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIF 1518
>CK605905
Length = 430
Score = 33.5 bits (75), Expect = 0.22
Identities = 30/107 (28%), Positives = 42/107 (39%), Gaps = 7/107 (6%)
Frame = +1
Query: 12 KEPTNV--PHCEDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVT 69
K P N P E I + K+PP E + + +NPGK P++
Sbjct: 43 KNPPNFFSPEGEKKIPHGPIGGGEKKKKTPPGPKNKGEGAPGKNPIFPQNPGKRGGPKIR 222
Query: 70 -PTPPKRSKSCPVTYKSRQATVKGKGKQKVVK----TPSEKKKRKVP 111
P P K+ K P ++ +KG K+ K PS KKK P
Sbjct: 223 GPPPSKKGKKKPPRGPPKKKNLKGGEKRSPKKGEIFAPSPKKKNGKP 363
>TC203388 homologue to UP|H2B_GOSHI (O22582) Histone H2B, complete
Length = 675
Score = 33.5 bits (75), Expect = 0.22
Identities = 29/123 (23%), Positives = 63/123 (50%), Gaps = 1/123 (0%)
Frame = +2
Query: 93 KGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQNVPVVPIDR 152
+ K ++V P +KK P + + +E + ++ V D P+EKK +GK++P+ +
Sbjct: 62 RSKHQIVMAPKAEKK---PAEKKPAE-EKKSTVGDKAPAEKKPKAGKKLPKEGGAGGEGK 229
Query: 153 VSFHLEESVQKWK-FVCKIRMAKVREVGPDVLECKDVIVLIEKAGLMKTIQNVGRCYEKL 211
++SV+ +K ++ K+ +++V P D+ + + G+M + + +EKL
Sbjct: 230 KKKRNKKSVETYKIYIFKV----LKQVHP------DIGISSKAMGIMNSF--INDIFEKL 373
Query: 212 VRE 214
+E
Sbjct: 374 AQE 382
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 32.7 bits (73), Expect = 0.38
Identities = 15/51 (29%), Positives = 24/51 (46%)
Frame = +2
Query: 38 SPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQA 88
+PP P TS + + +P K +T TP+ S+ CP T S ++
Sbjct: 359 APPPAPPATSTSPTSRPARSASPPKTTTAPSTPSTSNPSRKCPTTSPSSKS 511
>TC205401 similar to GB|AAP37850.1|30725656|BT008491 At4g26630 {Arabidopsis
thaliana;} , partial (16%)
Length = 912
Score = 32.3 bits (72), Expect = 0.50
Identities = 32/121 (26%), Positives = 47/121 (38%), Gaps = 3/121 (2%)
Frame = +3
Query: 30 NDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQAT 89
N SP K P P S S E SD E+P S + K+ + P S + T
Sbjct: 129 NKSRSPPKRAPKK-PSSNLSKSDEDSD-ESPKVFSRKKKNEKGGKQKTATPTKSASEEKT 302
Query: 90 VK---GKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQNVP 146
K GKGK+K PS+ + R ++ + A DI K++ P+
Sbjct: 303 EKVTRGKGKKKEKSRPSDNQLRDAICEILKEVNFNTATFTDILKKLAKQFDMDLTPRKAS 482
Query: 147 V 147
+
Sbjct: 483 I 485
>TC211694 weakly similar to UP|WAIP_HUMAN (O43516) Wiskott-Aldrich syndrome
protein interacting protein (WASP interacting protein)
(PRPL-2 protein), partial (5%)
Length = 782
Score = 32.0 bits (71), Expect = 0.65
Identities = 16/52 (30%), Positives = 22/52 (41%)
Frame = +1
Query: 60 PGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVP 111
P S+P +PTPP S P + + + G V TPS +R P
Sbjct: 463 PPPASSPRGSPTPPPTSAPAPFSSPAAASPSSASGAGPTVTTPSTPSRRASP 618
>CF922098
Length = 263
Score = 32.0 bits (71), Expect = 0.65
Identities = 20/49 (40%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Frame = +2
Query: 62 KVSTPEVTPTPPKRSKSCPVTYKSR-QATVKGKGKQKVVKTPSEKKKRK 109
K P TP KR K P K + + + G GKQK+ PS KKK+K
Sbjct: 2 KTKKP*KTP*KKKRPKKPPPPPKGKGKNPLGGGGKQKISHPPSPKKKKK 148
>TC227268
Length = 894
Score = 31.6 bits (70), Expect = 0.85
Identities = 39/176 (22%), Positives = 68/176 (38%), Gaps = 28/176 (15%)
Frame = +1
Query: 2 RKTYMSKVQNKEPTNVP------HCEDVT-------DEEEINDEDSPVK--------SPP 40
RK Y V+N P+ V H E VT E+E+ + +K S P
Sbjct: 325 RKVYNPVVENAVPSEVESEMPKDHLEKVTVTSCLAVSEQEVISSNEKIKKEAILTVSSVP 504
Query: 41 DVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGKGKQKVVK 100
D+ SVS+E+SD TP + +T K + +V + K + +
Sbjct: 505 DIETTPRLSVSKEVSD--------TPSSYQVTVESKPLTEITTKDKNISVSDEVKNEPID 660
Query: 101 TPSE--KKKRKVPVDVEDSESDAEADVQDIRPSEK-----KKYSGKRIPQNVPVVP 149
P K + + + S + + ++ +P++K K+ + QN P +P
Sbjct: 661 LPEPICKDENSHLTNGDLSHKEDQIGSENQKPNQKASIVAKQERAENGIQNSPTLP 828
>CF922867
Length = 450
Score = 31.6 bits (70), Expect = 0.85
Identities = 17/52 (32%), Positives = 24/52 (45%)
Frame = +1
Query: 35 PVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSR 86
P++ PP PG + + E +ENPG P P PP++ P K R
Sbjct: 226 PLEKPP---PGQKPELGPEPGPEENPGPKGPPGKPPWPPEKLPPDPGKEKPR 372
>AI443254 weakly similar to GP|9759050|dbj| AAA-type ATPase-like protein
{Arabidopsis thaliana}, partial (4%)
Length = 392
Score = 31.2 bits (69), Expect = 1.1
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -1
Query: 449 CFASQAARRRTTRR*ARSCCYCCSG 473
C +S ++ ++T R R CCYCC G
Sbjct: 227 CTSSGSSAQKTAFRTVRRCCYCCCG 153
>CF922271
Length = 442
Score = 31.2 bits (69), Expect = 1.1
Identities = 28/95 (29%), Positives = 42/95 (43%), Gaps = 18/95 (18%)
Frame = +2
Query: 35 PVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTP----TPPKRSKSCP----VTYKSR 86
P K P PG ++ + ++ GKV +P+ P TPPK KS P + +++R
Sbjct: 158 PQKKPRGKPPGGPPRARKK-NKKKRGGKVFSPKSPPGPPKTPPKGKKSPPRGVALFWETR 334
Query: 87 QATVKG----------KGKQKVVKTPSEKKKRKVP 111
+ KG KG ++ KTP KK P
Sbjct: 335 KGGKKGETPGKFLGN*KGGERTQKTPGGPKKGAPP 439
>TC227899
Length = 1311
Score = 31.2 bits (69), Expect = 1.1
Identities = 18/48 (37%), Positives = 24/48 (49%)
Frame = +3
Query: 30 NDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSK 77
ND P K V G+ SVS + ++Q + GK TPE T P +K
Sbjct: 555 NDTKEPEKEHK-VTTGISESVSADTTEQTDTGKPETPEAEITQPGITK 695
>CK606398
Length = 505
Score = 31.2 bits (69), Expect = 1.1
Identities = 30/127 (23%), Positives = 52/127 (40%)
Frame = +1
Query: 34 SPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQATVKGK 93
+P K+PP ++ ++E G+ V P PPK+ KS + ++ GK
Sbjct: 52 TPQKNPPP-------QTEKKTKNKEGVGRKKQKNVPPKPPKKRKSGTLKFRG------GK 192
Query: 94 GKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQNVPVVPIDRV 153
G +K+ P+ + + P + + ++ A R EKK G + P P R
Sbjct: 193 G-EKLKTPPNPQTRPSNPKRIREPQNRAPG-----RGPEKKFGGGGGQKKPPPKKPCQRT 354
Query: 154 SFHLEES 160
L E+
Sbjct: 355 *KRLPEN 375
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 30.8 bits (68), Expect = 1.5
Identities = 21/83 (25%), Positives = 35/83 (41%), Gaps = 5/83 (6%)
Frame = -2
Query: 35 PVKSPPDVVP-----GVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTYKSRQAT 89
PVK PP +P S S ++ + P V++P T TPP S+ P + A
Sbjct: 968 PVKLPPTTLPPTTPTATSPSSSTPLAVTQPPTVVASPPTTSTPPTTSQP-PANVAPKPAP 792
Query: 90 VKGKGKQKVVKTPSEKKKRKVPV 112
VK + + + ++P+
Sbjct: 791 VKPSAPKVAPASSPKAPPPQIPI 723
>TC215074 similar to UP|O04237 (O04237) Transcription factor, partial (88%)
Length = 1720
Score = 30.8 bits (68), Expect = 1.5
Identities = 30/129 (23%), Positives = 53/129 (40%), Gaps = 13/129 (10%)
Frame = +1
Query: 21 EDVTDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVST-PEVTPTPPKRSKSC 79
++V +E E N D DV G + S + E ++E K T E +R K+
Sbjct: 826 DEVANETEKNLGDEKPAVEEDVADGNKDSPTNETEEKEPEDKEMTLEEYEKVLEERRKAF 1005
Query: 80 PVT-----------YKSRQATVKGKGKQKV-VKTPSEKKKRKVPVDVEDSESDAEADVQD 127
++S QA K + +K S+K KRK + E+ + + +
Sbjct: 1006QALKTEERKVDTKEFESMQALSSKKDNHDIFIKLGSDKDKRKEAFEKEEKSKKSVSITEF 1185
Query: 128 IRPSEKKKY 136
++P+E + Y
Sbjct: 1186LKPAEGEAY 1212
>TC219933 weakly similar to UP|Q9EQF1 (Q9EQF1) GABA-A epsilon subunit splice
variant (Fragment), partial (7%)
Length = 490
Score = 30.8 bits (68), Expect = 1.5
Identities = 21/103 (20%), Positives = 42/103 (40%), Gaps = 1/103 (0%)
Frame = +1
Query: 19 HCEDVTDEEEINDEDSPVKSP-PDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSK 77
H E E + E +P P P P + +++ P S PE+ P P S+
Sbjct: 109 HSEMPEPESQNGHEPAPEPEPEPQPQPETKPEPTEQTQPHLEPKSESVPELEPNPQPESE 288
Query: 78 SCPVTYKSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESD 120
PV + QA ++ K + ++ +R+ + ++ ++
Sbjct: 289 --PVPTEQTQAPLEPKSGSEADPVVNDADRRETTIHSNEANAN 411
>TC204617 similar to UP|Q9ASZ9 (Q9ASZ9) At1g60730/F8A5_24, partial (98%)
Length = 1579
Score = 30.8 bits (68), Expect = 1.5
Identities = 12/26 (46%), Positives = 13/26 (49%)
Frame = +1
Query: 450 FASQAARRRTTRR*ARSCCYCCSGWE 475
F SQ RR R CC CC GW+
Sbjct: 1045 FVSQTNSRRHGRARVLCCCRCCQGWQ 1122
>BU927486 weakly similar to GP|20804751|dbj P0698A10.6 {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 445
Score = 30.4 bits (67), Expect = 1.9
Identities = 23/123 (18%), Positives = 49/123 (39%)
Frame = +2
Query: 24 TDEEEINDEDSPVKSPPDVVPGVETSVSQEISDQENPGKVSTPEVTPTPPKRSKSCPVTY 83
T +EE+ D PD V + ++ Q+N + T P++ K
Sbjct: 92 TTQEEVVVSDVAPAEKPDTTNVVVVPLEKDAQPQQNEEE--------TSPEKKKENVTET 247
Query: 84 KSRQATVKGKGKQKVVKTPSEKKKRKVPVDVEDSESDAEADVQDIRPSEKKKYSGKRIPQ 143
++ K G V ++ S K++ + D+ ++E A +++ + K+ +P
Sbjct: 248 ETESEVSKPSGDGNVPESGSFKEESTIVSDLPETEKKALQELKQLIQEALNKHEFSAVPT 427
Query: 144 NVP 146
+ P
Sbjct: 428 SNP 436
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.333 0.142 0.464
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,950,646
Number of Sequences: 63676
Number of extensions: 337664
Number of successful extensions: 4098
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 3933
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4064
length of query: 496
length of database: 12,639,632
effective HSP length: 101
effective length of query: 395
effective length of database: 6,208,356
effective search space: 2452300620
effective search space used: 2452300620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0032.7


