

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
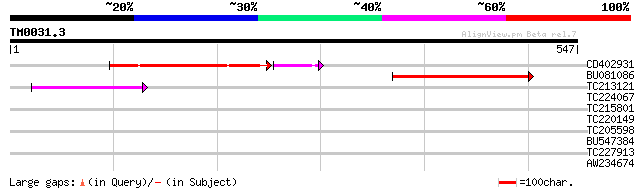
Query= TM0031.3
(547 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (ja... 159 2e-44
BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza s... 133 2e-31
TC213121 77 2e-14
TC224067 39 0.005
TC215801 similar to UP|HMAA_DROME (P29555) Homeobox protein abdo... 31 1.6
TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferas... 30 2.1
TC205598 similar to UP|BRU1_SOYBN (P35694) Brassinosteroid-regul... 30 3.6
BU547384 29 6.2
TC227913 similar to GB|AAP42720.1|30984514|BT008707 At4g26370 {A... 28 8.1
AW234674 weakly similar to GP|9294010|dbj| contains similarity t... 28 8.1
>CD402931 similar to GP|9081793|dbj P0489A01.23 {Oryza sativa (japonica
cultivar-group)}, partial (5%)
Length = 617
Score = 159 bits (403), Expect(2) = 2e-44
Identities = 89/156 (57%), Positives = 109/156 (69%)
Frame = +2
Query: 97 GTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLR 156
GT KTF+W L A +RS+GL+ + +ASLLLP TAHS FSIP+ I + STCN+
Sbjct: 11 GTSKTFLWKTLSAGMRSKGLMSSSCLKR-LASLLLPRG*TAHSTFSIPLVIKDDSTCNIN 187
Query: 157 QGSPKAELLKKASLIIWDEAPMLNKHCIEALDRSLNDIMKTQSTHGYDIPFGGKVVVLGD 216
QGS +++LL LIIWDE P++NK C EALDR+L DIM +Q+ PFGGK VVLG
Sbjct: 188 QGSARSKLLLHTKLIIWDETPVMNKFCFEALDRTLQDIMASQNKDNATKPFGGK-VVLGG 364
Query: 217 DFGQILPVISKGSRSEIVGSAINSSYLWKHCKVMKL 252
DF QILPVI KGSR +IVGSAIN+S KV+KL
Sbjct: 365 DFRQILPVIRKGSRQDIVGSAINASK-----KVLKL 457
Score = 38.5 bits (88), Expect(2) = 2e-44
Identities = 20/48 (41%), Positives = 27/48 (55%)
Frame = +1
Query: 255 NMSLQNATSTSSATEIKEFVDWLLQVGDGTVKTIDEEETLIEIPPDLL 302
NM L + +I+EFVDW+L + DG DE E I+IP +LL
Sbjct: 463 NMRLGTTNNNEDRDDIREFVDWILMIRDGNRDEEDEGE--IDIPKNLL 600
>BU081086 weakly similar to GP|27817864|db OJ1081_B12.13 {Oryza sativa
(japonica cultivar-group)}, partial (7%)
Length = 426
Score = 133 bits (334), Expect = 2e-31
Identities = 64/136 (47%), Positives = 86/136 (63%)
Frame = +2
Query: 370 KSDEDSGDNAE*FTSEVLNDFKCSEIPNHAIKLKAGVPIMLIRNIDQAAGLCNGTRMIVN 429
KSD + T+E +N S +PNH IKLK IML+RN+DQ GLCN TR+I+
Sbjct: 17 KSDTIESEGFSTITTEFINSLSTSRLPNHKIKLKVDSRIMLLRNLDQNEGLCNSTRLIIT 196
Query: 430 ALTKYIIVVTVLNENKMGETTFIPRMSLTPSNSDIPFKFQRRQFSVTLCFVMTINKSQGQ 489
+II +++ G T +IPR++ +PS S PFK RR+F + + + MTINKSQGQ
Sbjct: 197 MFADHIIEAKIMSGKGQGNTVYIPRLATSPSQSPWPFKLIRRKFPIIVSYAMTINKSQGQ 376
Query: 490 SLSHVGLYLPRPVFTH 505
L+ VGLYLP PVF+H
Sbjct: 377 LLASVGLYLPTPVFSH 424
>TC213121
Length = 713
Score = 77.0 bits (188), Expect = 2e-14
Identities = 40/112 (35%), Positives = 62/112 (54%)
Frame = +2
Query: 22 SLKEFPCLPYPKFSEIHNFEVIFVADELNYNRAEMVKIHDEFVSSLTSEQEIFYKNVLDD 81
S + FP + YP ++ + + +EL Y++ + SLT E + ++
Sbjct: 275 SHRNFPLMSYPIGYIVNPYGNKLIYNELAYDKDILAA*FSRCYHSLTDEHASIFNKIMHM 454
Query: 82 VLSDNS*FFFLYGFGGTGKTFVWNILYAALRSRGLIVLNVASSGIASLLLPG 133
+ + + +FLYG+GGTGKTFVW L + + S IVL +ASSGI S+LLPG
Sbjct: 455 IATQSGGVYFLYGYGGTGKTFVWKTLSSTIHSNSGIVLTMASSGILSMLLPG 610
>TC224067
Length = 415
Score = 39.3 bits (90), Expect = 0.005
Identities = 27/52 (51%), Positives = 34/52 (64%), Gaps = 2/52 (3%)
Frame = -2
Query: 466 FKFQRRQFSVTLCFVMTINKSQGQSLSHV-GLYLPRPV-FTHGQPYVALYRV 515
FKFQ + +C+ M INKS GQ+LS V G+ LPRP +HGQ YV L +V
Sbjct: 387 FKFQ--DVNSLVCYTMKINKSHGQTLSQVGGVLLPRPN**SHGQ-YVTLNQV 241
>TC215801 similar to UP|HMAA_DROME (P29555) Homeobox protein abdominal-A,
partial (3%)
Length = 654
Score = 30.8 bits (68), Expect = 1.6
Identities = 20/47 (42%), Positives = 25/47 (52%)
Frame = +2
Query: 457 LTPSNSDIPFKFQRRQFSVTLCFVMTINKSQGQSLSHVGLYLPRPVF 503
LTPSNS +PF F FS LC + S G+ S + L+ P VF
Sbjct: 329 LTPSNSSLPFTF---FFSFQLCPLGKKKSSMGKFSSLLLLFFPLLVF 460
>TC220149 weakly similar to UP|O23406 (O23406) Glucosyltransferase like
protein, partial (27%)
Length = 820
Score = 30.4 bits (67), Expect = 2.1
Identities = 12/37 (32%), Positives = 22/37 (59%)
Frame = -2
Query: 273 FVDWLLQVGDGTVKTIDEEETLIEIPPDLLIEQCKEP 309
FV W + G+ T+ T + + +++E P + +Q KEP
Sbjct: 360 FVPWSVHCGNATIGTPEPKLSIVEFHPQCVTKQPKEP 250
>TC205598 similar to UP|BRU1_SOYBN (P35694) Brassinosteroid-regulated protein
BRU1 precursor , partial (88%)
Length = 1015
Score = 29.6 bits (65), Expect = 3.6
Identities = 16/46 (34%), Positives = 27/46 (57%), Gaps = 1/46 (2%)
Frame = -1
Query: 227 KGSRSEIVGSAINSSYLWKHCKVMKLTVNMS-LQNATSTSSATEIK 271
K +RSE+ A+N + LW+ K++ + ++ L N + SSA IK
Sbjct: 886 KKNRSEVSAPALNRNALWESLKIVAVVIDHEVLLNPSQFSSAIGIK 749
>BU547384
Length = 537
Score = 28.9 bits (63), Expect = 6.2
Identities = 19/61 (31%), Positives = 31/61 (50%)
Frame = -2
Query: 109 AALRSRGLIVLNVASSGIASLLLPGDRTAHSRFSIPISINEISTCNLRQGSPKAELLKKA 168
+ALR + L LN+ S IA LL + + H P + ++T L GS E+ +++
Sbjct: 320 SALRLKPLFGLNIIDSYIAEKLLIDESSEHDNLGFPDVNSGMTTATLGMGS--GEIRRRS 147
Query: 169 S 169
S
Sbjct: 146 S 144
>TC227913 similar to GB|AAP42720.1|30984514|BT008707 At4g26370 {Arabidopsis
thaliana;} , partial (39%)
Length = 591
Score = 28.5 bits (62), Expect = 8.1
Identities = 12/32 (37%), Positives = 19/32 (58%)
Frame = -1
Query: 217 DFGQILPVISKGSRSEIVGSAINSSYLWKHCK 248
DFGQ +G R+ +VG N S++W+ C+
Sbjct: 249 DFGQ------RGRRTVVVGFVQNGSFVWRECE 172
>AW234674 weakly similar to GP|9294010|dbj| contains similarity to disease
resistance response protein~gene_id:MMM17.6 {Arabidopsis
thaliana}, partial (23%)
Length = 289
Score = 28.5 bits (62), Expect = 8.1
Identities = 11/23 (47%), Positives = 19/23 (81%)
Frame = -3
Query: 106 ILYAALRSRGLIVLNVASSGIAS 128
ILYA+ + +GL+ L VA++G++S
Sbjct: 278 ILYASRKDKGLVCLQVANAGVSS 210
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,560,506
Number of Sequences: 63676
Number of extensions: 292511
Number of successful extensions: 1355
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 1347
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1354
length of query: 547
length of database: 12,639,632
effective HSP length: 102
effective length of query: 445
effective length of database: 6,144,680
effective search space: 2734382600
effective search space used: 2734382600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0031.3


