

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
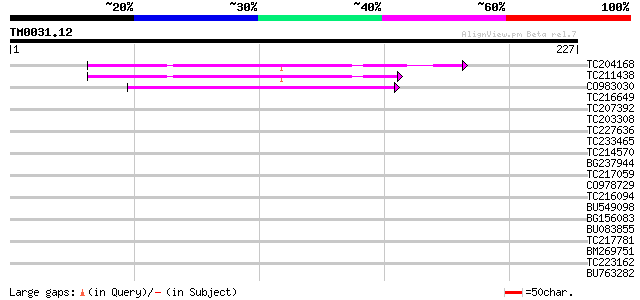
Query= TM0031.12
(227 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime ... 50 8e-07
TC211438 UP|GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain p... 50 1e-06
CO983030 46 1e-05
TC216649 weakly similar to UP|Q840B9 (Q840B9) Endo-b1,4-mannanas... 36 0.016
TC207392 similar to UP|BNK_DROME (P40794) Bottleneck protein, pa... 35 0.036
TC203308 35 0.036
TC227636 weakly similar to UP|Q9SYP0 (Q9SYP0) F9H16.4 protein, p... 34 0.048
TC233465 similar to UP|Q9VAQ7 (Q9VAQ7) CG1420-PA, partial (3%) 34 0.062
TC214570 homologue to PIR|T05766|T05766 peptidylprolyl isomerase... 34 0.062
BG237944 33 0.081
TC217059 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, parti... 32 0.24
CO978729 32 0.24
TC216094 similar to UP|Q6SJQ9 (Q6SJQ9) TFIID component TAF2 (Fra... 31 0.40
BU549098 31 0.53
BG156083 30 0.90
BU083855 30 1.2
TC217781 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecr... 29 1.5
BM269751 29 1.5
TC223162 29 2.0
BU763282 29 2.0
>TC204168 UP|Q7XXT2 (Q7XXT2) Prepro beta-conglycinin alpha prime subunit,
complete
Length = 2055
Score = 50.1 bits (118), Expect = 8e-07
Identities = 44/159 (27%), Positives = 69/159 (42%), Gaps = 7/159 (4%)
Frame = +3
Query: 32 QNTDLEKTEDQASNACKGNNNHKIQRNEQKYTRNSKINKITENAAPIITETTKKREENHK 91
+N K D N +GN ++ ++R + K + S ++ + A +I + R N
Sbjct: 258 KNAKKVKFHDHDHNTRRGNVSNTVRRRKTKVS--SHVHSHSHAHANLIKRKSTSRRRNTN 431
Query: 92 TLPERKHRASNESKAK-------TQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKS 144
+ RK+ E K + L K KRK+ N K T E+ VKKK KK+
Sbjct: 432 GIARRKNTEEREVKRNKMNVNTHAHTNLIKRKRKSTNGNTSRKSTKERKVKKK----KKT 599
Query: 145 KTIFTTSTERAKQRNRRITHGEVRFSLTEEENAERVRTR 183
KT + T+RAK+ L+ +EN E +R R
Sbjct: 600 KTRMRSKTKRAKKVK----------VLSLKENHEDIRIR 686
>TC211438 UP|GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor,
complete
Length = 1959
Score = 49.7 bits (117), Expect = 1e-06
Identities = 38/133 (28%), Positives = 60/133 (44%), Gaps = 7/133 (5%)
Frame = +2
Query: 32 QNTDLEKTEDQASNACKGNNNHKIQRNEQKYTRNSKINKITENAAPIITETTKKREENHK 91
+N K D N +GN ++ ++R + K + S ++ + A +I + R N
Sbjct: 236 KNAKKVKFHDHDHNTRRGNVSNTVRRRKTKVS--SHVHSHSHAHANLIKRKSTSRRRNTN 409
Query: 92 TLPERKHRASNESKAK-------TQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKS 144
+ RK+ E K + L K KRK+ N K T E+ VKKK KK+
Sbjct: 410 GIARRKNTEEREVKRNKMNVNTHAHTNLIKRKRKSTNGNTSRKSTKERKVKKK----KKT 577
Query: 145 KTIFTTSTERAKQ 157
KT + T+RAK+
Sbjct: 578 KTRMKSKTKRAKK 616
>CO983030
Length = 505
Score = 46.2 bits (108), Expect = 1e-05
Identities = 28/109 (25%), Positives = 51/109 (46%)
Frame = +3
Query: 48 KGNNNHKIQRNEQKYTRNSKINKITENAAPIITETTKKREENHKTLPERKHRASNESKAK 107
K K ++ ++K + K K + + KK+++ K ++K + + K K
Sbjct: 6 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 185
Query: 108 TQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAK 156
+ K KK K+K + + K+ +K KKK+K NKK K T++ K
Sbjct: 186 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKNKKKKXXKIKKTQKKK 332
>TC216649 weakly similar to UP|Q840B9 (Q840B9) Endo-b1,4-mannanase 26B,
partial (7%)
Length = 470
Score = 35.8 bits (81), Expect = 0.016
Identities = 23/96 (23%), Positives = 46/96 (46%), Gaps = 12/96 (12%)
Frame = -3
Query: 80 TETTKKREENHKTLPERKHRASNESKA-----------KTQSKLKKMKRKTQNAGQIMKQ 128
+E K++ ++HK ++ + ++ SKA K ++K+K+KT + + +
Sbjct: 318 SEEQKEKSKSHKRTSKKSVKENSVSKADRTSSAKKTPVKDAKSIEKIKKKTTSKKSVAEH 139
Query: 129 TSEKI-VKKKQKINKKSKTIFTTSTERAKQRNRRIT 163
S VK KQ +KK KT+ + K +++ T
Sbjct: 138 DSASASVKSKQPASKKQKTVSEKQDTKGKTASKKQT 31
>TC207392 similar to UP|BNK_DROME (P40794) Bottleneck protein, partial (7%)
Length = 1544
Score = 34.7 bits (78), Expect = 0.036
Identities = 45/199 (22%), Positives = 80/199 (39%), Gaps = 31/199 (15%)
Frame = +2
Query: 25 LFFFFP--FQNTDLEKTEDQASNACKGNNNHK-----IQRNEQKYTRNSKINKITENAAP 77
LFFF F E + + + G+N+ + ++++ + K K T+N
Sbjct: 65 LFFFVSVCFSGVCSEHQPESKALSSGGSNSRRRMLLEFDAEKEEFEQPPK-TKSTKNQTK 241
Query: 78 IITETTKKREENH-KTLPERKHRASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKI-VK 135
+I TT +N KT+ +++N + +K Q KL K K AG + T+E + VK
Sbjct: 242 LIKPTTNLSSKNQTKTI-----KSNNLNSSKNQLKLAKTKAA---AGDTLTNTTEVVAVK 397
Query: 136 KKQKINKKSKTIFTTS----------------------TERAKQRNRRITHGEVRFSLTE 173
K KSK + +TS A +++ +T+ +F
Sbjct: 398 KLNSTTLKSKKLNSTSKGLTKSSSLDLAKTSGGKNKTTKATATNKDKEVTNKVTKFLDQT 577
Query: 174 EENAERVRTRNPNQRENGG 192
E + + T N ++E G
Sbjct: 578 ESDKKSNNTNNKKKKEQQG 634
>TC203308
Length = 1778
Score = 34.7 bits (78), Expect = 0.036
Identities = 19/58 (32%), Positives = 30/58 (50%)
Frame = -3
Query: 103 ESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAKQRNR 160
+ K K + K KK K+K + + K+ +K KKK+K KK K ++ K +NR
Sbjct: 312 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKIFLGGPLKKTKFKNR 139
Score = 26.9 bits (58), Expect = 7.6
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = -2
Query: 112 LKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSK 145
LKK K+K + + K+ +K KKK+K KK K
Sbjct: 319 LKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 218
>TC227636 weakly similar to UP|Q9SYP0 (Q9SYP0) F9H16.4 protein, partial (8%)
Length = 1200
Score = 34.3 bits (77), Expect = 0.048
Identities = 28/125 (22%), Positives = 48/125 (38%), Gaps = 9/125 (7%)
Frame = +3
Query: 72 TENAAPIITETTKKREENHKTLPERKHRASNESKAKTQSKLKKMKR---------KTQNA 122
T+ +I + K R+E + + K R KAK + KK K Q
Sbjct: 417 TKEEEELILKAEKARKEEEEAKLKEKRRLEEIEKAKEALQRKKRNAEKAQQRAALKAQKE 596
Query: 123 GQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAKQRNRRITHGEVRFSLTEEENAERVRT 182
++ ++ EK KKK++ K S + +TE+ LTE+ +
Sbjct: 597 AELKEKEREKRAKKKER-RKTSSAVTAENTEQESAHTTETLTSVEESDLTEKPAEVTKKP 773
Query: 183 RNPNQ 187
+ P+Q
Sbjct: 774 QKPSQ 788
>TC233465 similar to UP|Q9VAQ7 (Q9VAQ7) CG1420-PA, partial (3%)
Length = 719
Score = 33.9 bits (76), Expect = 0.062
Identities = 19/52 (36%), Positives = 27/52 (51%)
Frame = -3
Query: 105 KAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAK 156
K K + K KK K+K + + K+ + KKK+K NKK KT T + K
Sbjct: 717 KKKKKKKKKKKKKKKKKKKKKKKKKKNQKKKKKKKKNKKKKTNSTPXXXKKK 562
Score = 30.0 bits (66), Expect = 0.90
Identities = 19/63 (30%), Positives = 28/63 (44%)
Frame = -2
Query: 97 KHRASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAK 156
K + + K K + K KK K+K K+ K KKK+K KK + T ++
Sbjct: 718 KKKKKKKKKKKKKKKKKKKKKK-------KKKKKPKKKKKKKKKQKKKNKLNTXXXQKKG 560
Query: 157 QRN 159
Q N
Sbjct: 559 QSN 551
>TC214570 homologue to PIR|T05766|T05766 peptidylprolyl isomerase M4E13.20 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(13%)
Length = 944
Score = 33.9 bits (76), Expect = 0.062
Identities = 21/72 (29%), Positives = 37/72 (51%)
Frame = +2
Query: 67 KINKITENAAPIITETTKKREENHKTLPERKHRASNESKAKTQSKLKKMKRKTQNAGQIM 126
K+ K+ E + E KK ++ K + A++ + KT+ K KK K K +++ +
Sbjct: 440 KVKKVDEETVEV--EVEKKEKKKKKKKNKENSEAASSDEEKTEKKKKKHKDKVEDSSPEL 613
Query: 127 KQTSEKIVKKKQ 138
+ SEK KKK+
Sbjct: 614 DK-SEKKKKKKK 646
>BG237944
Length = 486
Score = 33.5 bits (75), Expect = 0.081
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 2/86 (2%)
Frame = -3
Query: 118 KTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAKQRNRRITHGEVRFSLTEEENA 177
K N +I K S++ ++ +K +K KTI+ + E+ +++NRR + E E
Sbjct: 334 KRCNGAKISKL*SKQ--RRAEKNKRKKKTIYD*NEEKEEKKNRR------KLRKKEPE*R 179
Query: 178 ERVRTRNPNQR--ENGGVRENGNVNL 201
ERV+ RN +R E G R + ++ L
Sbjct: 178 ERVKNRNAVRR*IEEGSKRRDEDLTL 101
>TC217059 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, partial (9%)
Length = 1565
Score = 32.0 bits (71), Expect = 0.24
Identities = 25/119 (21%), Positives = 49/119 (41%)
Frame = +1
Query: 39 TEDQASNACKGNNNHKIQRNEQKYTRNSKINKITENAAPIITETTKKREENHKTLPERKH 98
+ED + +G + + +R+E+KYT + + + ++ E ++R+E K E K
Sbjct: 916 SEDSGKHRREGKDRKRHKRSEKKYTSSDE-DYSDDSELEDRKEAKRRRKEEKKLQKEEKR 1092
Query: 99 RASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAKQ 157
R E + + + + + +K+ KSKT + E AKQ
Sbjct: 1093 RRREEKRRRREER------------------------RAEKLKMKSKTDDISDDEEAKQ 1197
>CO978729
Length = 377
Score = 32.0 bits (71), Expect = 0.24
Identities = 21/55 (38%), Positives = 27/55 (48%)
Frame = +1
Query: 91 KTLPERKHRASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSK 145
K LP ESKA T KK K+K + + K+ +K KKK+K KK K
Sbjct: 52 KILPTLAIAYLEESKAFTCKGKKKKKKKKKKKKKKKKKPKKKKKKKKKKXXKKQK 216
Score = 29.3 bits (64), Expect = 1.5
Identities = 20/55 (36%), Positives = 25/55 (45%)
Frame = +2
Query: 97 KHRASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTS 151
KH E K K + K KK K+K ++ +K KKK NKK K T S
Sbjct: 95 KHLHVREKKKKKKKKKKKKKKKKN------QKKKKKKKKKKXXKNKKKKXKCTXS 241
>TC216094 similar to UP|Q6SJQ9 (Q6SJQ9) TFIID component TAF2 (Fragment),
partial (20%)
Length = 2400
Score = 31.2 bits (69), Expect = 0.40
Identities = 23/112 (20%), Positives = 48/112 (42%)
Frame = +3
Query: 40 EDQASNACKGNNNHKIQRNEQKYTRNSKINKITENAAPIITETTKKREENHKTLPERKHR 99
ED +S++ +NN + + + + P+ E + + +E HK+ +++ R
Sbjct: 1557 EDPSSSSIIQDNNIDADARRYASLQTLSVARFDPDGEPLGKEISARGKEKHKSKEKKRKR 1736
Query: 100 ASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTS 151
SN+ L++ + K + K+ EK + K Q K +I +S
Sbjct: 1737 ESNKGHHDDPEYLERKRLKKE------KKRREKELAKLQSDEAKRSSIDMSS 1874
>BU549098
Length = 571
Score = 30.8 bits (68), Expect = 0.53
Identities = 15/39 (38%), Positives = 20/39 (50%)
Frame = -1
Query: 181 RTRNPNQRENGGVRENGNVNLHSSLSYKSAVCVSVSDES 219
+ RNP RE GGV+E N+H + + S S ES
Sbjct: 154 KRRNPATREKGGVKEEA*QNIHEGACFGLSTSGSFSKES 38
>BG156083
Length = 372
Score = 30.0 bits (66), Expect = 0.90
Identities = 21/51 (41%), Positives = 24/51 (46%), Gaps = 15/51 (29%)
Frame = +2
Query: 87 EENHKTLPERKHRASNESKAKTQSK---------------LKKMKRKTQNA 122
EENH T E KH A+NES A +S KK K+KT NA
Sbjct: 68 EENHVTAMEEKHAAANES-AHAESNGVSHGADHGWQKVTYAKKQKKKTVNA 217
>BU083855
Length = 425
Score = 29.6 bits (65), Expect = 1.2
Identities = 21/60 (35%), Positives = 33/60 (55%), Gaps = 8/60 (13%)
Frame = -1
Query: 50 NNNHKIQRNEQKYTRNSKINKITENAAPIITETTKKREEN-----HKTLP---ERKHRAS 101
+N+H Q N K + +NK T++ +P+IT K R +N H+T P E KH+A+
Sbjct: 362 SNDHMKQLNSIK--DDILLNKTTKHFSPLITLNCKARADNKGNEIHQTEPLQKEAKHKAT 189
>TC217781 similar to UP|O81316 (O81316) F6N15.20 protein (Scarecrow-like 6)
(SCL6) (AT4g00150/F6N15_20), partial (42%)
Length = 1762
Score = 29.3 bits (64), Expect = 1.5
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 12/116 (10%)
Frame = +1
Query: 52 NHKIQRNEQKYTRNSKINKITENAAPIITETTKKREENHKTLPE----RKHRASNESKAK 107
+ K+Q ++ N ++ NA ++ T + +ENH+ LP+ R + N K
Sbjct: 67 DEKLQVLNPQFILNQNQSQFMPNAGLVLPLTYGQLQENHQLLPQPPAKRLNCGPNYQVPK 246
Query: 108 T-------QSKLKKMKRKTQNAGQIMKQTSEKIVKKKQK-INKKSKTIFTTSTERA 155
T + L++ +++ Q + Q +V KQK +N S+ + T ++A
Sbjct: 247 TPFLDSGQELLLRRQQQQLQLLPHHLLQRPSMVVAPKQKMVNSGSQDLATHQLQQA 414
>BM269751
Length = 394
Score = 29.3 bits (64), Expect = 1.5
Identities = 18/54 (33%), Positives = 26/54 (47%)
Frame = +3
Query: 90 HKTLPERKHRASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKK 143
H TL ++K + + K K + K KK K+K + KKK+K NKK
Sbjct: 75 HYTLLKKKKKKKKKKKKKKKKKKKKKKKKKKK-------------KKKKKKNKK 197
Score = 26.9 bits (58), Expect = 7.6
Identities = 14/34 (41%), Positives = 20/34 (58%)
Frame = +3
Query: 112 LKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSK 145
LKK K+K + + K+ +K KKK+K KK K
Sbjct: 87 LKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 188
Score = 26.6 bits (57), Expect = 9.9
Identities = 14/36 (38%), Positives = 21/36 (57%)
Frame = +1
Query: 109 QSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKS 144
+ K KK K+K + + K+ +K KKK+K KKS
Sbjct: 94 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKTKKS 201
>TC223162
Length = 782
Score = 28.9 bits (63), Expect = 2.0
Identities = 33/145 (22%), Positives = 60/145 (40%), Gaps = 7/145 (4%)
Frame = -2
Query: 51 NNHKIQRNEQKYTRNSKIN-KITENAAPIITETTKKREENHKTLPERKHRASNESKAKTQ 109
N H Q KY++NSK + K ++N R + L R RA ++S +
Sbjct: 766 NKHHQQNTSFKYSKNSKEHQKQSKN*TTYHQIFRVTRSKRLGKLFLR*PRAISQSNLSGR 587
Query: 110 SKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKSKTIFTTSTERAKQRNRRITHGEVRF 169
S KK+++ N+ ++ +Q ++ + + + K+S +I I H ++
Sbjct: 586 STHKKLEKVAVNSPRVTEQLAKPVPQGIRHTLKRSNSI-------------NILHQNLKA 446
Query: 170 SLTEEENAE------RVRTRNPNQR 188
L + N +R RNP R
Sbjct: 445 HLLQRRNRNINTCIPSLRIRNPLNR 371
>BU763282
Length = 421
Score = 28.9 bits (63), Expect = 2.0
Identities = 22/92 (23%), Positives = 42/92 (44%)
Frame = +3
Query: 85 KREENHKTLPERKHRASNESKAKTQSKLKKMKRKTQNAGQIMKQTSEKIVKKKQKINKKS 144
+RE+ ++ ++ R + + K + + KK KRK + + + K+ SEK K + +K S
Sbjct: 63 QREDEKRS---KRRRTVEDEERKQRKREKKEKRKDKKSHEHSKENSEKGKLKDKHRSKHS 233
Query: 145 KTIFTTSTERAKQRNRRITHGEVRFSLTEEEN 176
K + + + E L EE+N
Sbjct: 234 KVEGHMDFQELSNDDYFAKNNEFATWLKEEKN 329
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.307 0.122 0.323
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,880,073
Number of Sequences: 63676
Number of extensions: 85706
Number of successful extensions: 756
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 748
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 753
length of query: 227
length of database: 12,639,632
effective HSP length: 94
effective length of query: 133
effective length of database: 6,654,088
effective search space: 884993704
effective search space used: 884993704
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0031.12


