

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.10
(482 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
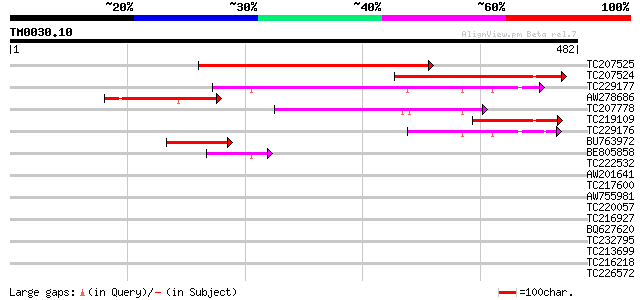
Score E
Sequences producing significant alignments: (bits) Value
TC207525 weakly similar to UP|NGP1_HUMAN (Q13823) Autoantigen NG... 379 e-105
TC207524 weakly similar to GB|AAH00107.1|12652715|BC000107 nucle... 207 1e-53
TC229177 weakly similar to UP|Q21086 (Q21086) C. elegans NST-1 p... 167 1e-41
AW278686 125 5e-29
TC207778 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (41%) 100 2e-21
TC219109 88 7e-18
TC229176 64 2e-10
BU763972 52 8e-07
BE805858 45 9e-05
TC222532 similar to UP|Q9BLS9 (Q9BLS9) MMR_HSR1 GTP-binding prot... 40 0.003
AW201641 39 0.007
TC217600 homologue to UP|NOG1_ARATH (Q9C6I8) Probable nucleolar ... 37 0.026
AW755981 37 0.026
TC220057 similar to GB|AAP41843.1|32396052|AY254472 short integu... 36 0.033
TC216927 homologue to UP|Q9SVA6 (Q9SVA6) GTP-binding-like protei... 35 0.097
BQ627620 similar to GP|10176988|db GTP-binding protein-like {Ara... 34 0.13
TC232795 similar to UP|Q9XI26 (Q9XI26) F9L1.38, partial (49%) 32 0.82
TC213699 similar to UP|Q9XI26 (Q9XI26) F9L1.38, partial (11%) 32 0.82
TC216218 homologue to UP|Q9M7P3 (Q9M7P3) GTP-binding protein, co... 31 1.4
TC226572 similar to UP|Q93W37 (Q93W37) AT5g01350/T10O8_60, parti... 30 2.4
>TC207525 weakly similar to UP|NGP1_HUMAN (Q13823) Autoantigen NGP-1, partial
(26%)
Length = 603
Score = 379 bits (972), Expect = e-105
Identities = 183/200 (91%), Positives = 192/200 (95%)
Frame = +2
Query: 161 LKENCKYKHMMLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQ 220
LKENCK+KHM+LLLNKCDL+PAWATKGWLRVLSKE+PTLAFHA+INKSFGKGSLLSVLRQ
Sbjct: 2 LKENCKHKHMVLLLNKCDLVPAWATKGWLRVLSKEFPTLAFHANINKSFGKGSLLSVLRQ 181
Query: 221 FARLKSDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLI 280
FARLK DKQAISVGF+GYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLI
Sbjct: 182 FARLKRDKQAISVGFVGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLI 361
Query: 281 DCPGVVYQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWENEND 340
DCPGVVY N D+E DVVLKGVVRVTNLKDAAD IGEVLKRV KEHL RAYKIKEW++END
Sbjct: 362 DCPGVVYHNNDTETDVVLKGVVRVTNLKDAADHIGEVLKRVKKEHLERAYKIKEWDDEND 541
Query: 341 FLLQLCKLTGKLLKGGEPDL 360
FLLQLCK TGKLLKGGEPDL
Sbjct: 542 FLLQLCKSTGKLLKGGEPDL 601
>TC207524 weakly similar to GB|AAH00107.1|12652715|BC000107 nucleolar GTPase
{Homo sapiens;} , partial (8%)
Length = 655
Score = 207 bits (526), Expect = 1e-53
Identities = 102/146 (69%), Positives = 124/146 (84%)
Frame = +2
Query: 328 RAYKIKEWENENDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPPQQED 387
RAYKIKEW++ENDFLLQLCK TGKL KGGEPDLMTAAKM+LHDWQRG+IPFFVPPP+QED
Sbjct: 2 RAYKIKEWDDENDFLLQLCKSTGKLXKGGEPDLMTAAKMILHDWQRGRIPFFVPPPRQED 181
Query: 388 LSEEPIVNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSVPVQKDLFTENELEEEADNLL 447
LSEEP VNG+D+DD VD N+ASAA+KAIA+VLSSQQQ +VPVQ DL++E EL+ EA +
Sbjct: 182 LSEEPNVNGVDLDDSVDGNEASAAMKAIADVLSSQQQINVPVQNDLYSEKELKGEATD-- 355
Query: 448 LISGDSPDGDVLDSDSDTSEQDPDTE 473
+ + ++L DS+TSE+DP +E
Sbjct: 356 QHPNTNANEEILALDSNTSEEDPISE 433
>TC229177 weakly similar to UP|Q21086 (Q21086) C. elegans NST-1 protein
(Corresponding sequence K01C8.9), partial (21%)
Length = 1353
Score = 167 bits (422), Expect = 1e-41
Identities = 105/299 (35%), Positives = 166/299 (55%), Gaps = 17/299 (5%)
Frame = +2
Query: 173 LLNKCDLIPAWATKGWLRVLSKEYPTLAFHAS-------INKSFGKGSLLSVLRQFARLK 225
LLNK DL+P A + WL+ L +E PT+AF S ++ G +L+ +L+ ++R
Sbjct: 2 LLNKIDLVPKEALEKWLKYLREELPTVAFKCSTQQQRSNLSDCLGADTLIKLLKNYSRSH 181
Query: 226 SDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGV 285
K++I+VG IG PNVGKSS+IN+L+ +V V PG T+ Q + L K + L+DCPGV
Sbjct: 182 EIKKSITVGLIGLPNVGKSSLINSLKRSHVVNVGSTPGLTRSMQEVQLDKNVKLLDCPGV 361
Query: 286 V-YQNKDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKIKEWE--NENDFL 342
V ++++++A V L R+ L D + E+ K E L YKI ++ + +DFL
Sbjct: 362 VMLKSQENDASVALLNCKRIEKLDDPISPVKEIFKLCPPEQLVTHYKIGTFKFGDVDDFL 541
Query: 343 LQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPP--QQEDLSEEPIVNGLDVD 400
L++ + GKL KGG D+ AA++VLHDW GKI ++ PP Q + +E IV+ +
Sbjct: 542 LKIATVRGKLKKGGIVDINAAARIVLHDWNEGKIIYYTIPPNRDQGEPAEVKIVSEFSKE 721
Query: 401 DGVDSNQAS-----AAIKAIANVLSSQQQSSVPVQKDLFTENELEEEADNLLLISGDSP 454
+D S ++K++ ++ + + SS P+ D E LE+E G+ P
Sbjct: 722 FNIDEVYKSESSYIGSLKSVDDLNAVEVPSSRPLNLD---ETMLEDETQIKPAEQGEGP 889
>AW278686
Length = 310
Score = 125 bits (313), Expect = 5e-29
Identities = 64/105 (60%), Positives = 82/105 (77%), Gaps = 5/105 (4%)
Frame = +2
Query: 81 KAHVAQEAFEEKYAAGASGTAEANEADGFIDLLGHTMFQKGQTKHISGELYKVIDSSDVV 140
KA +Q+AFEEK+ G+ +AEAN+ DGF DL+ HTM ++GQ+ + GELY VI SSDVV
Sbjct: 2 KADGSQDAFEEKH--GSRVSAEANDEDGFTDLVLHTMVEQGQSVRLWGELYTVIHSSDVV 175
Query: 141 VQ-----VLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLI 180
Q VLDARDP GTRCYHL+KHLK+NC+++ M+LLLNKCDL+
Sbjct: 176 DQAAESMVLDARDPPGTRCYHLKKHLKDNCQHERMVLLLNKCDLL 310
>TC207778 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (41%)
Length = 1511
Score = 100 bits (248), Expect = 2e-21
Identities = 62/196 (31%), Positives = 101/196 (50%), Gaps = 15/196 (7%)
Frame = +1
Query: 226 SDKQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGV 285
S + VGF+GYPNVGKSS IN L + V PG+TK +Q + ++ + L DCPG+
Sbjct: 298 SSSSNVIVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISDELTLCDCPGL 477
Query: 286 VYQN-KDSEADVVLKGVVRVTNLKDAADRIGEVLKRVMKEHLHRAYKI-----KEWENE- 338
V+ + S +++ GV+ + + + + + V RV + + YKI K +E++
Sbjct: 478 VFPSFSSSRYEMIACGVLPIDRMTEQRESVQVVADRVPRHVIEEIYKIRLPKPKSYESQS 657
Query: 339 -----NDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPP---QQEDLSE 390
++ L C G + G PD AA+++L D+ GK+P + PP +E E
Sbjct: 658 RPPLASELLRAYCTSRGYVASSGLPDETRAARLMLKDYTDGKLPHYQKPPGASDEEQALE 837
Query: 391 EPIVNGLDVDDGVDSN 406
EP+ + L D DS+
Sbjct: 838 EPVGHDLVDLDASDSS 885
>TC219109
Length = 732
Score = 88.2 bits (217), Expect = 7e-18
Identities = 46/77 (59%), Positives = 61/77 (78%)
Frame = +2
Query: 394 VNGLDVDDGVDSNQASAAIKAIANVLSSQQQSSVPVQKDLFTENELEEEADNLLLISGDS 453
VNG+D+DD VD N+ASAA+KAIA+VLSSQQQ+SVPVQ DL++ENEL+ EA + +
Sbjct: 2 VNGVDLDDSVDGNEASAAMKAIADVLSSQQQTSVPVQNDLYSENELKGEAAD--QYPNTN 175
Query: 454 PDGDVLDSDSDTSEQDP 470
+ D+L DS+TSE+DP
Sbjct: 176 ANEDILAPDSNTSEEDP 226
>TC229176
Length = 887
Score = 63.5 bits (153), Expect = 2e-10
Identities = 45/138 (32%), Positives = 71/138 (50%), Gaps = 7/138 (5%)
Frame = +3
Query: 339 NDFLLQLCKLTGKLLKGGEPDLMTAAKMVLHDWQRGKIPFFVPPP--QQEDLSEEPIVNG 396
+DFLL++ + GKL KGG D+ AA++VLHDW GKI ++ PP Q + SE IV+
Sbjct: 15 SDFLLKVGTVRGKLKKGGIVDINAAARIVLHDWNEGKIIYYTMPPNRDQGEPSEANIVSD 194
Query: 397 LDVDDGVDSNQAS-----AAIKAIANVLSSQQQSSVPVQKDLFTENELEEEADNLLLISG 451
+ +D +S ++ ++ N + + SS P+ D E LE+E G
Sbjct: 195 FSKEFNIDEVYSSESSYIGSLISVDNFNAVEVPSSHPLTLD---EMMLEDETQIKPAEQG 365
Query: 452 DSPDGDVLDSDSDTSEQD 469
+ P G++ + D E D
Sbjct: 366 EGP-GNLAEVDESMEEDD 416
>BU763972
Length = 409
Score = 51.6 bits (122), Expect = 8e-07
Identities = 23/56 (41%), Positives = 38/56 (67%)
Frame = +1
Query: 134 IDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWL 189
++ SD++V V+D+RDP RC LE + +E ++K +LL+NK DL+PA + W+
Sbjct: 1 VERSDLLVMVVDSRDPLFYRCPDLEAYAREVDEHKRTLLLVNKADLLPASVREKWV 168
>BE805858
Length = 300
Score = 44.7 bits (104), Expect = 9e-05
Identities = 22/63 (34%), Positives = 38/63 (59%), Gaps = 7/63 (11%)
Frame = +1
Query: 168 KHMMLLLNKCDLIPAWATKGWLRVLSKEYPTLAFHAS-------INKSFGKGSLLSVLRQ 220
+ ++LLLNK DL+P A + WL+ L +E PT+AF S ++ G +L+ +L+
Sbjct: 1 ERLVLLLNKIDLVPKEALEKWLKYLREELPTVAFKCSTQQQRSNLSDCLGADTLIKLLKN 180
Query: 221 FAR 223
++R
Sbjct: 181 YSR 189
>TC222532 similar to UP|Q9BLS9 (Q9BLS9) MMR_HSR1 GTP-binding protein,
partial (7%)
Length = 456
Score = 39.7 bits (91), Expect = 0.003
Identities = 23/43 (53%), Positives = 26/43 (59%), Gaps = 4/43 (9%)
Frame = +3
Query: 15 LSFSAMVKKKES----SGKSKQSLDANGNNDAKMEQSNSRVQR 53
LS SAM KKKE SGK K SLD N +NDAK E ++ R
Sbjct: 30 LSVSAMAKKKEKKVNVSGKPKHSLDVNRSNDAKKESRSAATVR 158
>AW201641
Length = 425
Score = 38.5 bits (88), Expect = 0.007
Identities = 30/128 (23%), Positives = 59/128 (45%), Gaps = 11/128 (8%)
Frame = +3
Query: 134 IDSSDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL- 192
++ S V++ ++D + + L++N K+++L +NKC+ R++
Sbjct: 57 VEESSVIIFLVDGQAGLTAADEEIADWLRKNYSDKYVILAVNKCE-------SPRKRIMQ 215
Query: 193 SKEYPTLAFHA---SINKSFGKGSLLSV-------LRQFARLKSDKQAISVGFIGYPNVG 242
+ E+ +L F S G G LL + + + L + ++ +G PNVG
Sbjct: 216 ASEFWSLGFEPLPISAISGTGTGELLDLVCSGLQKIEESNNLDEEDYVPAISIVGRPNVG 395
Query: 243 KSSVINTL 250
KSS++N L
Sbjct: 396 KSSILNAL 419
>TC217600 homologue to UP|NOG1_ARATH (Q9C6I8) Probable nucleolar GTP-binding
protein 1, partial (31%)
Length = 630
Score = 36.6 bits (83), Expect = 0.026
Identities = 39/145 (26%), Positives = 63/145 (42%), Gaps = 8/145 (5%)
Frame = +2
Query: 174 LNKCDLIPAWATKGWLRVLSKEYPTLAFHASINKSFGKGSLLSVLRQFARLKS-DKQAIS 232
L +C + A V+ + P+LA+ L V + ARL S D +
Sbjct: 170 LYRCKCLKVAALGRMCTVIKRVGPSLAY------------LEQVRQHMARLPSIDPNTRT 313
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKV-------WQYITLTKRIFLIDCPGV 285
V GYPNVGKSS IN + +V V P TK ++Y+ R +ID PG+
Sbjct: 314 VLICGYPNVGKSSFINKITRADV-DVQPYAFTTKSLFVGHTDYKYL----RYQVIDTPGI 478
Query: 286 VYQNKDSEADVVLKGVVRVTNLKDA 310
+ + + + + + + +L+ A
Sbjct: 479 LDRPFEDRNIIEMCSITALAHLRAA 553
>AW755981
Length = 451
Score = 36.6 bits (83), Expect = 0.026
Identities = 20/53 (37%), Positives = 29/53 (53%), Gaps = 1/53 (1%)
Frame = +3
Query: 233 VGFIGYPNVGKSSVINTL-RTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPG 284
+ F G NVGKSS++N L R V + + PG T+ + L + L+D PG
Sbjct: 117 IAFAGRSNVGKSSLLNALTRQWGVVRTSDKPGLTQTINFFNLGTKHCLVDLPG 275
>TC220057 similar to GB|AAP41843.1|32396052|AY254472 short integuments 2
{Arabidopsis thaliana;} , partial (35%)
Length = 556
Score = 36.2 bits (82), Expect = 0.033
Identities = 34/116 (29%), Positives = 54/116 (46%), Gaps = 2/116 (1%)
Frame = +2
Query: 137 SDVVVQVLDARDPQGTRCYHLEKHLKENCKYKHMMLLLNKCDLIPAWATKGWLRVL-SKE 195
+D+V++V DAR P + L+ HL K ++ LNK DL W S
Sbjct: 200 ADLVIEVRDARIPFSSANADLQPHLSA----KRRVVALNKKDLANPNIMHKWTHYFESCN 367
Query: 196 YPTLAFHASINKSFGKGSLLSVLR-QFARLKSDKQAISVGFIGYPNVGKSSVINTL 250
+A +A S K LL V+ + + + + V +G PNVGKS+++N +
Sbjct: 368 QNCVAINAHSMSSVKK--LLEVVEFKLKEVICREPTLLVMVVGVPNVGKSALMNCI 529
>TC216927 homologue to UP|Q9SVA6 (Q9SVA6) GTP-binding-like protein, complete
Length = 1771
Score = 34.7 bits (78), Expect = 0.097
Identities = 21/71 (29%), Positives = 36/71 (50%), Gaps = 10/71 (14%)
Frame = +2
Query: 233 VGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLT----------KRIFLIDC 282
VG +G+P+VGKS+++N K+ E +++ TLT +I L+D
Sbjct: 407 VGLVGFPSVGKSTLLN--------KLTGTFSEVASYEFTTLTCIPGVITYRGAKIQLLDL 562
Query: 283 PGVVYQNKDSE 293
PG++ KD +
Sbjct: 563 PGIIEGAKDGK 595
>BQ627620 similar to GP|10176988|db GTP-binding protein-like {Arabidopsis
thaliana}, partial (21%)
Length = 431
Score = 34.3 bits (77), Expect = 0.13
Identities = 20/69 (28%), Positives = 36/69 (51%), Gaps = 3/69 (4%)
Frame = +1
Query: 228 KQAISVGFIGYPNVGKSSVINTLRTKNVCKVAPIPGETK---VWQYITLTKRIFLIDCPG 284
K + + +G PNVGKS+++N L ++ V P G T+ Q+ + I+L+D G
Sbjct: 112 KLPLQLAIVGRPNVGKSTLLNALLQEDRVLVGPEAGLTRDSIRTQFEFQGRTIYLVDTAG 291
Query: 285 VVYQNKDSE 293
+ + K +
Sbjct: 292 WLQRTKQEK 318
>TC232795 similar to UP|Q9XI26 (Q9XI26) F9L1.38, partial (49%)
Length = 1027
Score = 31.6 bits (70), Expect = 0.82
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = +2
Query: 437 NELEEEADNLLLISGDSPDGDVLDSDSDTSEQDPDTEVPYEQPHSA 482
+E E+E + + +I D D + DT E DPD + P EQ H+A
Sbjct: 629 DEEEDEGEMVPVIYEDKDSSD--EESDDTMETDPDEQQPEEQFHAA 760
>TC213699 similar to UP|Q9XI26 (Q9XI26) F9L1.38, partial (11%)
Length = 822
Score = 31.6 bits (70), Expect = 0.82
Identities = 17/46 (36%), Positives = 25/46 (53%)
Frame = +3
Query: 437 NELEEEADNLLLISGDSPDGDVLDSDSDTSEQDPDTEVPYEQPHSA 482
+E E+E + + +I D D + DT E DPD + P EQ H+A
Sbjct: 651 DEEEDEGEMVPVIYEDKDSSD--EESDDTMETDPDEQQPEEQFHAA 782
>TC216218 homologue to UP|Q9M7P3 (Q9M7P3) GTP-binding protein, complete
Length = 1545
Score = 30.8 bits (68), Expect = 1.4
Identities = 11/20 (55%), Positives = 16/20 (80%)
Frame = +3
Query: 231 ISVGFIGYPNVGKSSVINTL 250
+ +G +G PNVGKS++ NTL
Sbjct: 144 LKIGIVGLPNVGKSTLFNTL 203
>TC226572 similar to UP|Q93W37 (Q93W37) AT5g01350/T10O8_60, partial (57%)
Length = 704
Score = 30.0 bits (66), Expect = 2.4
Identities = 15/52 (28%), Positives = 25/52 (47%)
Frame = -1
Query: 54 PFTPVFGPKTKRKRPCLLAFDYQSLAKKAHVAQEAFEEKYAAGASGTAEANE 105
P PVF +K K PC+ F+ S K +E ++ + A+ A+ N+
Sbjct: 515 PPNPVFSSPSKLKNPCIPTFEITS*EPKYITTKEIYKHPALSPATSLAKENQ 360
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,195,890
Number of Sequences: 63676
Number of extensions: 249685
Number of successful extensions: 996
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 987
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 989
length of query: 482
length of database: 12,639,632
effective HSP length: 101
effective length of query: 381
effective length of database: 6,208,356
effective search space: 2365383636
effective search space used: 2365383636
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0030.10


