

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.6
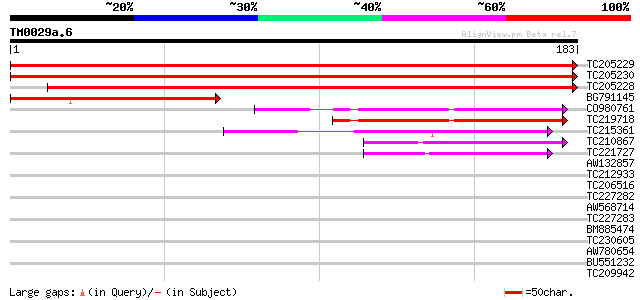
(183 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205229 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, par... 356 4e-99
TC205230 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, par... 355 7e-99
TC205228 317 2e-87
BG791145 130 3e-31
CO980761 62 2e-10
TC219718 weakly similar to UP|Q9LS39 (Q9LS39) Arabidopsis thalia... 57 6e-09
TC215361 48 2e-06
TC210867 weakly similar to UP|Q8VYU4 (Q8VYU4) At5g11350/F2I11_24... 46 1e-05
TC221727 similar to UP|Q9LS39 (Q9LS39) Arabidopsis thaliana geno... 41 4e-04
AW132857 similar to GP|17979010|gb At5g11350/F2I11_240 {Arabidop... 35 0.026
TC212933 similar to UP|Q8VYU4 (Q8VYU4) At5g11350/F2I11_240, part... 32 0.22
TC206516 similar to UP|Q9LIR6 (Q9LIR6) Beta-amylase (AT3g23920/F... 28 2.4
TC227282 homologue to GB|AAD14482.1|4249385|T2K10 T2K10.11 {Arab... 28 3.2
AW568714 homologue to GP|4249385|gb|A T2K10.11 {Arabidopsis thal... 28 3.2
TC227283 homologue to GB|AAD14482.1|4249385|T2K10 T2K10.11 {Arab... 27 5.4
BM885474 27 5.4
TC230605 homologue to UP|Q8QGI9 (Q8QGI9) CC chemokine with stalk... 27 5.4
AW780654 27 5.4
BU551232 27 7.0
TC209942 similar to UP|Q8S4Q6 (Q8S4Q6) Phytochrome A specific si... 26 9.2
>TC205229 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, partial (57%)
Length = 1743
Score = 356 bits (913), Expect = 4e-99
Identities = 173/183 (94%), Positives = 180/183 (97%)
Frame = +1
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNVH DLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNS PGSAPHALLAMGKV
Sbjct: 529 ANTHVNVHHDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSIPGSAPHALLAMGKV 708
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTN 120
DPSHPDLA+DPL+ILRPHSKLVHQLPLVSAYSSFART+GLG+EQHK RLD+ATNEPLFTN
Sbjct: 709 DPSHPDLAVDPLNILRPHSKLVHQLPLVSAYSSFARTVGLGFEQHKGRLDNATNEPLFTN 888
Query: 121 VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 180
VTRDFIG+LDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIA+LAEFRCCKN
Sbjct: 889 VTRDFIGSLDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIAMLAEFRCCKN 1068
Query: 181 RSR 183
RSR
Sbjct: 1069RSR 1077
>TC205230 similar to UP|Q8W0Z9 (Q8W0Z9) AT3g58560/F14P22_150, partial (48%)
Length = 1367
Score = 355 bits (911), Expect = 7e-99
Identities = 173/183 (94%), Positives = 178/183 (96%)
Frame = +3
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNV QDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNS PGSAPHALLAMGKV
Sbjct: 366 ANTHVNVSQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSVPGSAPHALLAMGKV 545
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTN 120
DPSHPDLA+DPL+ILRPHSKLVHQLPLVSAYSSFART+GLGYEQHKRR+D TNEPLFTN
Sbjct: 546 DPSHPDLAVDPLNILRPHSKLVHQLPLVSAYSSFARTVGLGYEQHKRRMDGGTNEPLFTN 725
Query: 121 VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 180
VTRDFIG LDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN
Sbjct: 726 VTRDFIGTLDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 905
Query: 181 RSR 183
+SR
Sbjct: 906 KSR 914
>TC205228
Length = 795
Score = 317 bits (812), Expect = 2e-87
Identities = 156/171 (91%), Positives = 161/171 (93%)
Frame = +1
Query: 13 DVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPL 72
DV QVH GLEKIAASADIPMLVCGDFNS PGSAPHALLAMGKVDPSHPDLA+DPL
Sbjct: 13 DVXXXQVHXXXXGLEKIAASADIPMLVCGDFNSIPGSAPHALLAMGKVDPSHPDLAVDPL 192
Query: 73 SILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYI 132
+ILRPHSKLVHQLPLVSAYSSFART+GLG+EQHKRRLD TNEPLFTNVTRDFIG+LDYI
Sbjct: 193 NILRPHSKLVHQLPLVSAYSSFARTVGLGFEQHKRRLDDTTNEPLFTNVTRDFIGSLDYI 372
Query: 133 FYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKNRSR 183
FYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKNRSR
Sbjct: 373 FYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKNRSR 525
>BG791145
Length = 401
Score = 130 bits (328), Expect = 3e-31
Identities = 66/76 (86%), Positives = 66/76 (86%), Gaps = 8/76 (10%)
Frame = +2
Query: 1 ANTHVNVHQDLKDVKLWQ--------VHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPH 52
ANTHVNVH DLKDVKLWQ VHTLLKGLEKIAASADIPMLVCGDFNS PGSAPH
Sbjct: 173 ANTHVNVHHDLKDVKLWQL*ISPSF*VHTLLKGLEKIAASADIPMLVCGDFNSIPGSAPH 352
Query: 53 ALLAMGKVDPSHPDLA 68
ALLAMGKVDPSHPDLA
Sbjct: 353 ALLAMGKVDPSHPDLA 400
>CO980761
Length = 877
Score = 61.6 bits (148), Expect = 2e-10
Identities = 38/101 (37%), Positives = 55/101 (53%)
Frame = -2
Query: 80 KLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSL 139
+L HQL L SAYS H+ R D EPL T+ F+G +DYI+++ D +
Sbjct: 714 RLQHQLKLCSAYSGVPGN-------HRTRDD--IGEPLATSYHSKFMGTVDYIWHSEDLI 562
Query: 140 VVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 180
V +LE L ++LR+ LPS +W SDH+A++ E N
Sbjct: 561 PVR-VLETLPIDTLRRSRGLPSEKWGSDHLAVVCEVAFANN 442
>TC219718 weakly similar to UP|Q9LS39 (Q9LS39) Arabidopsis thaliana genomic
DNA, chromosome 3, P1 clone: MYF24, partial (15%)
Length = 447
Score = 56.6 bits (135), Expect = 6e-09
Identities = 30/76 (39%), Positives = 46/76 (60%)
Frame = +1
Query: 105 HKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEW 164
H+ R D EPL T+ F+G +DYI+++ D + V +LE L ++LR+ LPS +W
Sbjct: 19 HRTRDD--IGEPLATSYHSKFMGTVDYIWHSEDLIPVR-VLETLPIDTLRRSRGLPSEKW 189
Query: 165 SSDHIALLAEFRCCKN 180
SDH+A++ EF N
Sbjct: 190 GSDHLAVVCEFAFANN 237
>TC215361
Length = 824
Score = 48.1 bits (113), Expect = 2e-06
Identities = 36/107 (33%), Positives = 49/107 (45%), Gaps = 1/107 (0%)
Frame = +2
Query: 70 DPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGAL 129
+P S L P +PL S Y+S EP FTN T DF G L
Sbjct: 170 NPSSNLMPDCLEESPIPLCSVYAS------------------TRGEPPFTNYTPDFTGTL 295
Query: 130 DYIFYT-ADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEF 175
DYI ++ +D + S LEL D ++ LP+ + SDH+ + AEF
Sbjct: 296 DYILFSPSDHIKPISFLELPDSDAADIVGGLPNFIYPSDHLPIGAEF 436
>TC210867 weakly similar to UP|Q8VYU4 (Q8VYU4) At5g11350/F2I11_240, partial
(16%)
Length = 733
Score = 45.8 bits (107), Expect = 1e-05
Identities = 22/66 (33%), Positives = 38/66 (57%)
Frame = +3
Query: 115 EPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAE 174
EPL T+ R F+G +DYI + ++ L +L + + +++ P+ +W SDHIAL+ E
Sbjct: 192 EPLVTSYNRRFLGTVDYI-WRSEGLQTTRVLAPISKHAMQWTPGFPTKKWGSDHIALVTE 368
Query: 175 FRCCKN 180
K+
Sbjct: 369 LAFLKD 386
>TC221727 similar to UP|Q9LS39 (Q9LS39) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MYF24, partial (32%)
Length = 953
Score = 40.8 bits (94), Expect = 4e-04
Identities = 21/61 (34%), Positives = 36/61 (58%)
Frame = +2
Query: 115 EPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAE 174
E L T+ F+G +DY++Y +D +V +L+ + L + LP + SDH+AL++E
Sbjct: 431 ELLATSYHSKFLGTVDYLWY-SDGIVPSRVLDTVSISDLLRTGGLPCKKVGSDHLALVSE 607
Query: 175 F 175
F
Sbjct: 608 F 610
>AW132857 similar to GP|17979010|gb At5g11350/F2I11_240 {Arabidopsis
thaliana}, partial (17%)
Length = 421
Score = 34.7 bits (78), Expect = 0.026
Identities = 18/49 (36%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Frame = +3
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-ADIPMLVCGDFNSNPGS 49
N HV + + ++KL QV LL + ++ D P+ +CGDFN P S
Sbjct: 252 NIHVLYNPNRGEIKLGQVRVLLDKAKAVSKLWNDAPVAICGDFNCTPKS 398
>TC212933 similar to UP|Q8VYU4 (Q8VYU4) At5g11350/F2I11_240, partial (7%)
Length = 512
Score = 31.6 bits (70), Expect = 0.22
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Frame = +1
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-ADIPMLVCGDFN 44
N HV + + ++KL QV LL + ++ D P+ +CGDFN
Sbjct: 376 NIHVLYNPNRGEIKLGQVRVLLDKAKAVSKLWNDAPVAICGDFN 507
>TC206516 similar to UP|Q9LIR6 (Q9LIR6) Beta-amylase (AT3g23920/F14O13_11),
partial (25%)
Length = 469
Score = 28.1 bits (61), Expect = 2.4
Identities = 19/60 (31%), Positives = 27/60 (44%), Gaps = 8/60 (13%)
Frame = -1
Query: 78 HSKLVHQ--------LPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGAL 129
H+ L HQ +PL SA+ RT+ + Q + SA EPL + +R G L
Sbjct: 385 HTSLFHQSHHQHPTVMPL*SAHEHTHRTVAAHFHQQVEPVPSALPEPLASQASRVHKGHL 206
>TC227282 homologue to GB|AAD14482.1|4249385|T2K10 T2K10.11 {Arabidopsis
thaliana;} , partial (61%)
Length = 1408
Score = 27.7 bits (60), Expect = 3.2
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +1
Query: 156 DTALPSPEWSSDHIALLAEFRCCKNRS 182
D P P W DH + + RCC RS
Sbjct: 565 DLGSPVPVWYKDHSSYCCKRRCCSIRS 645
>AW568714 homologue to GP|4249385|gb|A T2K10.11 {Arabidopsis thaliana},
partial (8%)
Length = 393
Score = 27.7 bits (60), Expect = 3.2
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +1
Query: 156 DTALPSPEWSSDHIALLAEFRCCKNRS 182
D P P W DH + + RCC RS
Sbjct: 247 DLGSPVPVWYKDHSSYCCKRRCCSIRS 327
>TC227283 homologue to GB|AAD14482.1|4249385|T2K10 T2K10.11 {Arabidopsis
thaliana;} , partial (27%)
Length = 1192
Score = 26.9 bits (58), Expect = 5.4
Identities = 11/27 (40%), Positives = 13/27 (47%)
Frame = +1
Query: 156 DTALPSPEWSSDHIALLAEFRCCKNRS 182
D P P W DH + + RCC RS
Sbjct: 46 DLGSPVPVWYKDHSSHCCKRRCCSIRS 126
>BM885474
Length = 420
Score = 26.9 bits (58), Expect = 5.4
Identities = 18/61 (29%), Positives = 27/61 (43%), Gaps = 14/61 (22%)
Frame = +3
Query: 39 VCGDFNSNP------------GSAPHALLAMGKVDPSHPDLAIDPLSILRPH--SKLVHQ 84
+CGD ++N S L M ++ P H + A+ L L PH SKL+ Q
Sbjct: 174 LCGDASNNSHSQ*LRS*RHIHNSQDRITLQMWRIPPKHAETALSALLSLMPHSSSKLLSQ 353
Query: 85 L 85
+
Sbjct: 354 V 356
>TC230605 homologue to UP|Q8QGI9 (Q8QGI9) CC chemokine with stalk CK2,
partial (6%)
Length = 659
Score = 26.9 bits (58), Expect = 5.4
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Frame = +3
Query: 131 YIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHI-ALLAEFRCCKNRSR 183
YI+Y ADSL++E +++ KD + E D ALL +R ++R R
Sbjct: 126 YIYYYADSLIME------NKKRKGKDYEVEDEEMKMDKFYALLRRYRDARDRRR 269
>AW780654
Length = 355
Score = 26.9 bits (58), Expect = 5.4
Identities = 8/19 (42%), Positives = 11/19 (57%)
Frame = +3
Query: 160 PSPEWSSDHIALLAEFRCC 178
P P WS H++ + F CC
Sbjct: 177 PPPSWSGSHLSRILFFSCC 233
>BU551232
Length = 545
Score = 26.6 bits (57), Expect = 7.0
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = +1
Query: 44 NSNPGSAPHA-----LLAMGKVDPSHPDLAIDPLSILRPHSKLVHQ 84
N +P P A L+ GK SH DPLS ++PH+ L HQ
Sbjct: 196 NIHPPQLPSAIVF*KLVIHGKTVFSHQ--LYDPLSEMKPHAHLSHQ 327
>TC209942 similar to UP|Q8S4Q6 (Q8S4Q6) Phytochrome A specific signal
transduction component PAT3, partial (13%)
Length = 594
Score = 26.2 bits (56), Expect = 9.2
Identities = 15/36 (41%), Positives = 22/36 (60%), Gaps = 3/36 (8%)
Frame = -1
Query: 139 LVVESLLELLDEES---LRKDTALPSPEWSSDHIAL 171
LVV SL+ELLD S ++ T SP WS +++ +
Sbjct: 327 LVVLSLIELLDLHSIFQMKGHTH*DSPHWSKENLRI 220
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,670,591
Number of Sequences: 63676
Number of extensions: 108729
Number of successful extensions: 492
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 486
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 486
length of query: 183
length of database: 12,639,632
effective HSP length: 91
effective length of query: 92
effective length of database: 6,845,116
effective search space: 629750672
effective search space used: 629750672
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0029a.6


