

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.5
(1389 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
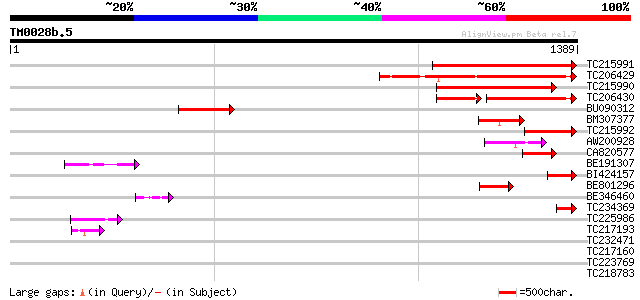
Score E
Sequences producing significant alignments: (bits) Value
TC215991 578 e-165
TC206429 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulator... 499 e-141
TC215990 470 e-132
TC206430 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulator... 257 3e-97
BU090312 214 3e-55
BM307377 191 3e-48
TC215992 189 1e-47
AW200928 107 4e-23
CA820577 weakly similar to PIR|T51447|T51 transcription regulato... 106 6e-23
BE191307 102 1e-21
BI424157 similar to PIR|T51447|T51 transcription regulator-like ... 93 9e-19
BE801296 89 2e-17
BE346460 62 1e-09
TC234369 UP|P93525 (P93525) PSbaNS5 protein, partial (5%) 57 4e-08
TC225986 weakly similar to UP|Q9W1I6 (Q9W1I6) CG18426-PA (LD1203... 49 2e-05
TC217193 similar to UP|Q7XJN2 (Q7XJN2) At2g40650 protein, partia... 44 5e-04
TC232471 similar to UP|NLFE_HUMAN (P18615) Negative elongation f... 42 0.002
TC217160 similar to UP|Q15410 (Q15410) Nucleic acid binding prot... 40 0.005
TC223769 38 0.035
TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%) 37 0.046
>TC215991
Length = 1356
Score = 578 bits (1489), Expect = e-165
Identities = 290/353 (82%), Positives = 314/353 (88%), Gaps = 1/353 (0%)
Frame = +1
Query: 1037 CSREEHEDREHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHE 1096
CSREEHED EHDNKAESEGEAEG+ DAHDVEGDGMSL SERFL TVKPLAK+VP LHE
Sbjct: 1 CSREEHEDGEHDNKAESEGEAEGIADAHDVEGDGMSLPYSERFLLTVKPLAKHVPPMLHE 180
Query: 1097 KDRNSQVFYGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNA 1156
KDRNS+VFYG+DS YVL RLHQTLYERIQSAK NSSSA++KWKAS DT STDQYDRFMNA
Sbjct: 181 KDRNSRVFYGNDSIYVLLRLHQTLYERIQSAKINSSSADRKWKASSDTSSTDQYDRFMNA 360
Query: 1157 LYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYA 1216
LYSLLDGSSDNTKFEDDCRAIIG QSY+LFTLDKLIYKLVKQLQA+A++EMD KLLQLYA
Sbjct: 361 LYSLLDGSSDNTKFEDDCRAIIGIQSYVLFTLDKLIYKLVKQLQAVAADEMDTKLLQLYA 540
Query: 1217 YEKSRKLGKFIDIVYHENACVILHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVD 1276
YEKSRK GKF+D+VYHENA V+LH+ENIYRIEYSPGP KLSIQLMD GHDKPEVTAVS+D
Sbjct: 541 YEKSRKPGKFVDMVYHENARVLLHDENIYRIEYSPGPMKLSIQLMDSGHDKPEVTAVSMD 720
Query: 1277 PKFSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFSSQAMEGLQVFNGLECKIACS 1336
P FSTYLHNDFLSVVPDKK+KS +FLKRNK RYA + EFSSQAMEGLQ+ NGLECKIACS
Sbjct: 721 PNFSTYLHNDFLSVVPDKKQKSGIFLKRNKRRYASNDEFSSQAMEGLQIINGLECKIACS 900
Query: 1337 SSKVSYVLDTEDVLYRKKSKRRILH-PTSSCNEPAKSSNIRSSRALRFCKLFS 1388
SSKVSYVLDTED L+R + KRR L +S +E A+SSNI SSR RF LFS
Sbjct: 901 SSKVSYVLDTEDFLFRIRRKRRALRLKSSGAHEQAQSSNICSSRVQRFRNLFS 1059
>TC206429 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulatory-like
protein, partial (23%)
Length = 1771
Score = 499 bits (1286), Expect = e-141
Identities = 284/493 (57%), Positives = 341/493 (68%), Gaps = 11/493 (2%)
Frame = +1
Query: 907 QGGDSTRPGTFTNGATTGGTEVHRYQEQSIRHFKSEREEGELSPNGDMEEDNFAVYENSG 966
+G D P NG + ++V + E S K E+EEGELSPNGD EEDN Y +S
Sbjct: 124 EGCDMATPVPVANGVLSESSKVKTHDE-SFGPCKIEKEEGELSPNGDSEEDNIVAYGDSN 300
Query: 967 LDAGHKGKGGDMSQKYQSRHGEQVCC-EAKGENDVDADDEGEESPHRSSDDSENASE-NV 1024
+ + K K +KYQSR+GE C EA G+ND DADDE DSEN SE
Sbjct: 301 VQSMAKSKHNIERRKYQSRNGEDESCPEAGGDNDADADDE----------DSENVSEAGE 450
Query: 1025 DVSGSESADGEECSREEHEDREH------DNKAESEGEAEGMVDAHDVEGDGMSLTISER 1078
DVSGSESA G+EC RE+HE+ E D KAESEGEAEG+ DA GDG SL +SER
Sbjct: 451 DVSGSESA-GDECFREDHEEEEDIEHDDVDGKAESEGEAEGICDAQ-AGGDGTSLPLSER 624
Query: 1079 FLQTVKPLAKYVPA-ALHEKDRNSQVFYGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKK 1137
FL +VKPL K+V A + E+ ++S+VFYG+D FYVLFRLHQ LYERI SAK +S SAE K
Sbjct: 625 FLSSVKPLTKHVSAVSFVEEMKDSRVFYGNDDFYVLFRLHQALYERILSAKTHSMSAEMK 804
Query: 1138 WKASHDTGSTDQYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVK 1197
WKA D S D Y RF+NALY+LLDGS++N KFED+CRAIIG QSY+LFTLDKLIYKLV+
Sbjct: 805 WKAK-DASSPDPYSRFINALYNLLDGSAENAKFEDECRAIIGNQSYVLFTLDKLIYKLVR 981
Query: 1198 QLQAIASEEMDNKLLQLYAYEKSRKLGKFIDIVYHENACVILHEENIYRIEYSPGPKKLS 1257
QLQ +A++E+DNKLLQLY YEKSRK GK D VYH NA VILHEENIYR++ S P +LS
Sbjct: 982 QLQTVATDEVDNKLLQLYEYEKSRKPGKLNDSVYHANAHVILHEENIYRLQCSSTPSRLS 1161
Query: 1258 IQLMDCGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFSS 1317
IQLMD ++KPE+ AVS+DP FS YLHNDFLSV P+KKE + L RNK +Y E S+
Sbjct: 1162 IQLMDNMNEKPELFAVSIDPNFSFYLHNDFLSVFPNKKEPHGIILHRNKRQYGKLDELSA 1341
Query: 1318 --QAMEGLQVFNGLECKIACSSSKVSYVLDTEDVLYRKKSKRRILHPTSSCNEPAKSSNI 1375
AMEG++V NGLECKIACSSSK+SYVLDT+D +R + KRR T+ +S
Sbjct: 1342 ICSAMEGVKVINGLECKIACSSSKISYVLDTQDFFFRPRKKRRTPSGTT-------TSRF 1500
Query: 1376 RSSRALRFCKLFS 1388
R R RF KL +
Sbjct: 1501 RRDREERFRKLLA 1539
>TC215990
Length = 884
Score = 470 bits (1209), Expect = e-132
Identities = 232/294 (78%), Positives = 262/294 (88%), Gaps = 1/294 (0%)
Frame = +2
Query: 1046 EHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPAALHEKDRNSQVFY 1105
EHDNKAESEGEAEGM DA+DVEGDG SL SERFL TVKPLAK+VP LH+K R +VFY
Sbjct: 2 EHDNKAESEGEAEGMTDANDVEGDGASLPYSERFLVTVKPLAKHVPPVLHDKQRTVRVFY 181
Query: 1106 GSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRFMNALYSLLDGSS 1165
G+DSFYVLFRLHQTLYERIQSAK NSSSAEKKW+AS+DTGS+DQY RFM+ALY+LLDGSS
Sbjct: 182 GNDSFYVLFRLHQTLYERIQSAKVNSSSAEKKWRASNDTGSSDQYGRFMDALYNLLDGSS 361
Query: 1166 DNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKLGK 1225
D+TKFED+CRAIIGTQSY+LFTLDKLIYKLVKQLQ +A+EEMDNKLLQLY YE SRK G+
Sbjct: 362 DSTKFEDECRAIIGTQSYVLFTLDKLIYKLVKQLQVVATEEMDNKLLQLYTYENSRKPGR 541
Query: 1226 FIDIVYHENACVILHEENIYRIEYSPGPKKL-SIQLMDCGHDKPEVTAVSVDPKFSTYLH 1284
F+D+VYHENA V+LH+ENIYRIE SP P +L SIQLMD G+DKPE+TAVS+DP FS YLH
Sbjct: 542 FVDLVYHENARVLLHDENIYRIECSPAPTQLSSIQLMDYGYDKPEMTAVSMDPNFSAYLH 721
Query: 1285 NDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFSSQAMEGLQVFNGLECKIACSSS 1338
NDFLSVVPDKKEKS ++LKRNK +YA S E+SSQ ++GL NGLECKIACSSS
Sbjct: 722 NDFLSVVPDKKEKSGIYLKRNKRKYAISDEYSSQTLDGLXXINGLECKIACSSS 883
>TC206430 similar to UP|Q9LFQ3 (Q9LFQ3) Transcriptional regulatory-like
protein, partial (14%)
Length = 1282
Score = 257 bits (656), Expect(2) = 3e-97
Identities = 135/222 (60%), Positives = 164/222 (73%), Gaps = 2/222 (0%)
Frame = +3
Query: 1169 KFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKLGKFID 1228
KFED+CRAIIG QSY+LFTLDKLIYKLV+QLQ +A++E+DNKLLQLY YEKSRK GK D
Sbjct: 324 KFEDECRAIIGNQSYVLFTLDKLIYKLVRQLQTVATDEVDNKLLQLYEYEKSRKSGKLND 503
Query: 1229 IVYHENACVILHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVDPKFSTYLHNDFL 1288
VYH NA VILHE+NIYR++ S P +L IQLMD ++KPE+ AVS+DP FS YLH+DFL
Sbjct: 504 SVYHANAHVILHEDNIYRLQCSSTPSRLFIQLMDNMNEKPELFAVSIDPNFSFYLHSDFL 683
Query: 1289 SVVPDKKEKSRLFLKRNKHRYAGSSEFSS--QAMEGLQVFNGLECKIACSSSKVSYVLDT 1346
SV P+KKE + L RNK +Y E S+ AMEG++V NGLECKIACSSSK+SYVLDT
Sbjct: 684 SVFPNKKEPHGIILHRNKRQYGNLDELSAICSAMEGVKVVNGLECKIACSSSKISYVLDT 863
Query: 1347 EDVLYRKKSKRRILHPTSSCNEPAKSSNIRSSRALRFCKLFS 1388
+D +R + KRR T+ +S R R RF KL +
Sbjct: 864 QDFFFRPRKKRRTPSGTT-------TSQSRRDREERFRKLLA 968
Score = 118 bits (296), Expect(2) = 3e-97
Identities = 68/113 (60%), Positives = 80/113 (70%), Gaps = 2/113 (1%)
Frame = +1
Query: 1046 EHDNKAESEGEAEGMVDAHDVEGDGMSLTISERFLQTVKPLAKYVPA-ALHEKDRNSQVF 1104
E D KAESEGEAEG+ DA GDG SL +SERFL +VKPL K+V A + E+ ++S+VF
Sbjct: 10 EFDGKAESEGEAEGICDAQ-AGGDGTSLPLSERFLSSVKPLTKHVSAVSFVEEMKDSRVF 186
Query: 1105 YGSDSFYVLFRLHQTLYERIQSAKFNSSSAEKKWKASHDTGSTDQYDRF-MNA 1156
YG+D FYV FRLHQ LYER+ SAK +S SAE KWKA D S D Y MNA
Sbjct: 187 YGNDDFYVFFRLHQALYERLLSAKTHSMSAEMKWKAK-DASSPDPYSSLKMNA 342
>BU090312
Length = 409
Score = 214 bits (544), Expect = 3e-55
Identities = 99/135 (73%), Positives = 116/135 (85%)
Frame = +1
Query: 415 SNCDRCTPSYRLLPKNYPIPIASYKSKIGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEES 474
S+C RCTPSYRLLP +YPIP AS +S++GA+VLNDHWVSVTSGSEDYSFKHMR+NQYEES
Sbjct: 1 SDCKRCTPSYRLLPADYPIPTASQRSELGAQVLNDHWVSVTSGSEDYSFKHMRRNQYEES 180
Query: 475 LFRCEDDRFELDMLLESVNLTTKRVEELFEKINNNVIKGDRPVRIDEHLTALNLRCIERL 534
LFRCEDDR+ELDMLLESV+ K+ EEL+ IN N I + RI++H T LNLRCIERL
Sbjct: 181 LFRCEDDRYELDMLLESVSSAAKKAEELYNSINENKISVETLNRIEDHFTVLNLRCIERL 360
Query: 535 YGDHGLDVMEVLRKN 549
YGDHGLDV+++LRKN
Sbjct: 361 YGDHGLDVIDILRKN 405
>BM307377
Length = 426
Score = 191 bits (484), Expect = 3e-48
Identities = 100/141 (70%), Positives = 108/141 (75%), Gaps = 28/141 (19%)
Frame = +3
Query: 1149 QYDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQ---------- 1198
QYDRFMNALYSLLDGSSDNTKFEDDCRAIIG QSY+LFTLDKLIYKLVKQ
Sbjct: 3 QYDRFMNALYSLLDGSSDNTKFEDDCRAIIGIQSYVLFTLDKLIYKLVKQVKYISSSFCL 182
Query: 1199 ------------------LQAIASEEMDNKLLQLYAYEKSRKLGKFIDIVYHENACVILH 1240
LQA+A++EMD KLLQLYAYEKSRK GKF+D+VYHENA V+LH
Sbjct: 183 LHTIISFLRLKYLFILF*LQAVAADEMDTKLLQLYAYEKSRKPGKFVDMVYHENARVLLH 362
Query: 1241 EENIYRIEYSPGPKKLSIQLM 1261
+ENIYRIEYSPGP KLSIQLM
Sbjct: 363 DENIYRIEYSPGPMKLSIQLM 425
>TC215992
Length = 1071
Score = 189 bits (479), Expect = 1e-47
Identities = 95/127 (74%), Positives = 103/127 (80%)
Frame = +3
Query: 1262 DCGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFSSQAME 1321
D GHDKPEVTAVS+DP FSTYLHNDFLSVVPDKKEKS +FLKRNK RYAG+ EFSSQAME
Sbjct: 3 DSGHDKPEVTAVSMDPNFSTYLHNDFLSVVPDKKEKSGIFLKRNKRRYAGNDEFSSQAME 182
Query: 1322 GLQVFNGLECKIACSSSKVSYVLDTEDVLYRKKSKRRILHPTSSCNEPAKSSNIRSSRAL 1381
GLQ+ NGLECKIACSSSKVSYVLDTED L+R + K+R+LHP SS S SSR
Sbjct: 183 GLQIINGLECKIACSSSKVSYVLDTEDFLFRIRRKKRVLHPKSSGAHEQAQSPKSSSRVQ 362
Query: 1382 RFCKLFS 1388
RF LFS
Sbjct: 363 RFRNLFS 383
>AW200928
Length = 457
Score = 107 bits (267), Expect = 4e-23
Identities = 65/156 (41%), Positives = 90/156 (57%), Gaps = 5/156 (3%)
Frame = +2
Query: 1164 SSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDNKLLQLYAYEKSRKL 1223
SSDNT+FEDD II Y +FT+ K IY+LVK Q +A++E D KL QLYAY+KSR
Sbjct: 2 SSDNTEFEDDRPPIIAIPPYDIFTIYKPIYQLVKHAQPVAADETDYKLSQLYAYDKSRTP 181
Query: 1224 GKFIDIVYHENACVI-----LHEENIYRIEYSPGPKKLSIQLMDCGHDKPEVTAVSVDPK 1278
+F DI YH N + + +I+ + P + +++ VS+DP
Sbjct: 182 AEFADIPYHRNDRALSMTRTYNRLHIHLTNETAYPTDGPLHMINL-----R*LHVSLDP* 346
Query: 1279 FSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSE 1314
FSTY+H DFLSV+ D K+ + LKR K R+A + E
Sbjct: 347 FSTYIHYDFLSVLSDTKQTPGILLKRTKRRHARNDE 454
>CA820577 weakly similar to PIR|T51447|T51 transcription regulator-like protein
- Arabidopsis thaliana, partial (6%)
Length = 480
Score = 106 bits (265), Expect = 6e-23
Identities = 55/86 (63%), Positives = 65/86 (74%), Gaps = 2/86 (2%)
Frame = +3
Query: 1257 SIQLMDCGHDKPEVTAVSVDPKFSTYLHNDFLSVVPDKKEKSRLFLKRNKHRYAGSSEFS 1316
SIQLMD ++KPE+ AVS+DP FS YLHNDFLSV P+KKE + L RNK +Y E S
Sbjct: 3 SIQLMDNMNEKPELFAVSIDPNFSFYLHNDFLSVFPNKKEPHGIILHRNKRQYGKLDELS 182
Query: 1317 S--QAMEGLQVFNGLECKIACSSSKV 1340
+ AMEG++V NGLECKIACSSSKV
Sbjct: 183 AIXSAMEGVKVINGLECKIACSSSKV 260
>BE191307
Length = 474
Score = 102 bits (254), Expect = 1e-21
Identities = 67/190 (35%), Positives = 95/190 (49%), Gaps = 6/190 (3%)
Frame = -2
Query: 135 DRRSAMPTVRQAHVVK----RERTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKE 190
+R S P +RQ K R+R D D S +RP+ D D+ ++ KEQR+R +E
Sbjct: 461 ERGSMAPMIRQMPADKQRYRRDRLPSHDRDHDMSAERPELDDDKTMMNIHKEQRKRESRE 282
Query: 191 KDHREDGKRDWERDGRDHEHDGGQ--YRERLSHKRKSDRKAEDFGAEPLLDADEFFGTRP 248
+ R+ +D R+H+ D + +R K+KS +KAE
Sbjct: 281 RRMRD-------QDEREHDLDNNRDLNLQRFPDKKKSVKKAEGM---------------- 171
Query: 249 MSSTCDDKKSLKTKYPEEFAFCDKVKEKLRNPDGYQEFLKCLHIYSKEIITQQELQSLVG 308
Y + F+FC+KVKEKL + D YQ FLKCLHI+S II + +L +LV
Sbjct: 170 --------------YSQAFSFCEKVKEKLSSSDDYQTFLKCLHIFSNGIIKRNDLPNLVT 33
Query: 309 DLLGKYPDLM 318
DLLGK+ DLM
Sbjct: 32 DLLGKHSDLM 3
>BI424157 similar to PIR|T51447|T51 transcription regulator-like protein -
Arabidopsis thaliana, partial (2%)
Length = 421
Score = 92.8 bits (229), Expect = 9e-19
Identities = 46/71 (64%), Positives = 55/71 (76%)
Frame = +1
Query: 1318 QAMEGLQVFNGLECKIACSSSKVSYVLDTEDVLYRKKSKRRILHPTSSCNEPAKSSNIRS 1377
Q ++GLQ+ NGLECKIACSSSKVSYVLDTED L++ + KRR L+ +SSC+ KS I S
Sbjct: 1 QTLDGLQIINGLECKIACSSSKVSYVLDTEDFLHQTRRKRRTLYQSSSCHGQEKSPIICS 180
Query: 1378 SRALRFCKLFS 1388
SR R CKLFS
Sbjct: 181 SRVQRSCKLFS 213
>BE801296
Length = 256
Score = 88.6 bits (218), Expect = 2e-17
Identities = 45/84 (53%), Positives = 60/84 (70%)
Frame = +2
Query: 1150 YDRFMNALYSLLDGSSDNTKFEDDCRAIIGTQSYLLFTLDKLIYKLVKQLQAIASEEMDN 1209
Y M+ALY+ L G D+TK+ED C A++ T SY LFTL L+YKL KQL A+A+E++D+
Sbjct: 5 YGMLMHALYNTLYGPHDSTKWEDHCYAMMRTYSYDLFTLYHLMYKLEKQLPAVATEDIDD 184
Query: 1210 KLLQLYAYEKSRKLGKFIDIVYHE 1233
KLL LYAYE R+ + +D YHE
Sbjct: 185 KLLHLYAYENPRQPVR*VDSYYHE 256
>BE346460
Length = 329
Score = 62.4 bits (150), Expect = 1e-09
Identities = 41/92 (44%), Positives = 54/92 (58%)
Frame = -3
Query: 309 DLLGKYPDLMEEFNEFLVQAEKNDGGFLAGVMNKKSLWIEGPGLKPMKAEDRDRDRDRDR 368
D K+PDLM+ FNE + Q +K MN+ S E K +K ED +RDR D
Sbjct: 288 DFPRKFPDLMKGFNECVTQCKK---------MNESSR-DEEHVQKQIKLEDWNRDRHHDG 139
Query: 369 YRDDGMKERDRECRERDRSTVVSNKEVSGLKI 400
DD +KE +CR+RD+STVV+NK VSG K+
Sbjct: 138 --DDRVKETVSDCRKRDKSTVVANKNVSGRKL 49
>TC234369 UP|P93525 (P93525) PSbaNS5 protein, partial (5%)
Length = 613
Score = 57.4 bits (137), Expect = 4e-08
Identities = 30/49 (61%), Positives = 36/49 (73%)
Frame = +1
Query: 1340 VSYVLDTEDVLYRKKSKRRILHPTSSCNEPAKSSNIRSSRALRFCKLFS 1388
VSYVLDTED L++ + KRR L+ +SSC+ KS I SSRA R CKLFS
Sbjct: 190 VSYVLDTEDFLHQTRRKRRTLYQSSSCHGQEKSPIICSSRAQRSCKLFS 336
>TC225986 weakly similar to UP|Q9W1I6 (Q9W1I6) CG18426-PA (LD12035p), partial
(13%)
Length = 1553
Score = 48.5 bits (114), Expect = 2e-05
Identities = 34/127 (26%), Positives = 63/127 (48%)
Frame = +2
Query: 150 KRERTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHE 209
+R+R ++D ++R + + +R +R E+EQRRRVE+ + E +DWE R+ E
Sbjct: 17 ERDRDRELKREKDRELERYEREAERERVRKEREQRRRVEEAERQYETYLKDWEYREREKE 196
Query: 210 HDGGQYRERLSHKRKSDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEEFAF 269
+ +E+ + + RK + E D DE R S ++K+ + + E+
Sbjct: 197 KERQYEKEKEKERERKRRKEILYDEE---DEDEDSRKRWRRSVIEEKRKKRLREKED-DL 364
Query: 270 CDKVKEK 276
D+ KE+
Sbjct: 365 VDRQKEE 385
>TC217193 similar to UP|Q7XJN2 (Q7XJN2) At2g40650 protein, partial (88%)
Length = 1691
Score = 43.9 bits (102), Expect = 5e-04
Identities = 34/91 (37%), Positives = 41/91 (44%), Gaps = 10/91 (10%)
Frame = +3
Query: 151 RERTTVSHCDRDPSVDRPDPDHDRGLLRAEKE--------QRRRVEKEKDH--REDGKRD 200
RER DRD DR D DR LR EK+ +R R EK++D R R
Sbjct: 954 RERGRGRDRDRDRERDR---DRDRYRLREEKDYGRDREGRERERREKDRDRGRRRSHSRS 1124
Query: 201 WERDGRDHEHDGGQYRERLSHKRKSDRKAED 231
R EHDGG YR+R + S R+ D
Sbjct: 1125 RSRSRDRREHDGGDYRKRHARSSVSPRRHGD 1217
Score = 36.6 bits (83), Expect = 0.079
Identities = 19/38 (50%), Positives = 24/38 (63%)
Frame = +3
Query: 349 GPGLKPMKAEDRDRDRDRDRYRDDGMKERDRECRERDR 386
G G + +RDRDRDR R R++ RDRE RER+R
Sbjct: 963 GRGRDRDRDRERDRDRDRYRLREEKDYGRDREGRERER 1076
Score = 32.0 bits (71), Expect = 1.9
Identities = 18/28 (64%), Positives = 21/28 (74%)
Frame = +3
Query: 359 DRDRDRDRDRYRDDGMKERDRECRERDR 386
DRD DRD DR R G ++RDR+ RERDR
Sbjct: 927 DRDYDRDYDRERGRG-RDRDRD-RERDR 1004
Score = 30.8 bits (68), Expect = 4.3
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 8/75 (10%)
Frame = +3
Query: 169 DPDHDRGLLRAEKEQRRRVEKEKDHRE-DGKRDWERD-------GRDHEHDGGQYRERLS 220
+ DH RG + R R HR+ D RD++RD GRD + D + R+R
Sbjct: 837 EKDHYRGRSPTRERDRDRRHDSHRHRDRDYDRDYDRDYDRERGRGRDRDRDRERDRDR-- 1010
Query: 221 HKRKSDRKAEDFGAE 235
R R+ +D+G +
Sbjct: 1011-DRYRLREEKDYGRD 1052
Score = 30.4 bits (67), Expect = 5.7
Identities = 17/28 (60%), Positives = 21/28 (74%)
Frame = +3
Query: 359 DRDRDRDRDRYRDDGMKERDRECRERDR 386
DRD DR+R R RD ++RDRE R+RDR
Sbjct: 939 DRDYDRERGRGRD---RDRDRE-RDRDR 1010
>TC232471 similar to UP|NLFE_HUMAN (P18615) Negative elongation factor E
(NELF-E) (RD protein), partial (11%)
Length = 480
Score = 41.6 bits (96), Expect = 0.002
Identities = 23/48 (47%), Positives = 29/48 (59%), Gaps = 6/48 (12%)
Frame = +3
Query: 353 KPMKAEDRDRDRDRDRYRD-DGMKERDRE-----CRERDRSTVVSNKE 394
KP DRDR+RDRD +RD D ++RDRE R+RDR N+E
Sbjct: 282 KPSSDRDRDRERDRDHHRDRDRDRDRDRERERERARDRDREAERRNRE 425
Score = 39.3 bits (90), Expect = 0.012
Identities = 27/88 (30%), Positives = 43/88 (48%)
Frame = +3
Query: 144 RQAHVVKRERTTVSHCDRDPSVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWER 203
RQ V ++R R+ S D P P DR R E+++DH D RD +R
Sbjct: 210 RQEEVADQKRRQEQLLSRNHSSD-PKPSSDRD---------RDRERDRDHHRDRDRDRDR 359
Query: 204 DGRDHEHDGGQYRERLSHKRKSDRKAED 231
D R+ E + + R+R + +R +R+ E+
Sbjct: 360 D-RERERERARDRDREAERRNREREREE 440
Score = 30.4 bits (67), Expect = 5.7
Identities = 13/32 (40%), Positives = 21/32 (65%)
Frame = +3
Query: 356 KAEDRDRDRDRDRYRDDGMKERDRECRERDRS 387
+ +R RDRDR+ R + +ER+ E R R+R+
Sbjct: 369 RERERARDRDREAERRNREREREEESRARERA 464
>TC217160 similar to UP|Q15410 (Q15410) Nucleic acid binding protein
(Fragment), partial (37%)
Length = 988
Score = 40.4 bits (93), Expect = 0.005
Identities = 21/44 (47%), Positives = 27/44 (60%), Gaps = 1/44 (2%)
Frame = +3
Query: 354 PMKAEDRDRDRDRDRYRD-DGMKERDRECRERDRSTVVSNKEVS 396
P DRDRDRDRDR RD + + RD++ R+RDR S + S
Sbjct: 21 PSTMTDRDRDRDRDRTRDRERERRRDKDDRDRDRDRARSKRSRS 152
>TC223769
Length = 674
Score = 37.7 bits (86), Expect = 0.035
Identities = 31/103 (30%), Positives = 47/103 (45%)
Frame = +1
Query: 164 SVDRPDPDHDRGLLRAEKEQRRRVEKEKDHREDGKRDWERDGRDHEHDGGQYRERLSHKR 223
S++R HDR +EKE R R +KD +++ +R + EH QYR +
Sbjct: 49 SLERSGRYHDRS--SSEKEHRYRSSTDKDKHSQKRKESDRSSSEKEH---QYRSSTDKDK 213
Query: 224 KSDRKAEDFGAEPLLDADEFFGTRPMSSTCDDKKSLKTKYPEE 266
S ++ E + P ++ G R SST DK K K +E
Sbjct: 214 HSRKRKESDRSSP----EKEHGLR--SSTDKDKHHRKRKERDE 324
>TC218783 similar to UP|Q40363 (Q40363) NuM1 protein, partial (19%)
Length = 870
Score = 37.4 bits (85), Expect = 0.046
Identities = 36/125 (28%), Positives = 51/125 (40%), Gaps = 1/125 (0%)
Frame = +3
Query: 940 KSEREEGELSPNGDMEEDNFAVYENSGLDAGHKGKGGDMSQKYQSRHGEQVCCEAKGEND 999
K E + E S + D EDN A + A K + S + E KG+ D
Sbjct: 432 KVESSDDESSESSD--EDNDA---KPAVTAVSKPSARAQKKVESSDSDDSSSDEDKGKMD 596
Query: 1000 VDADDEGEESPHRSSDDSENASENVDVSGS-ESADGEECSREEHEDREHDNKAESEGEAE 1058
+D DD+ SSD+SE + V S ES+D E E+ + + + G
Sbjct: 597 IDDDDD-------SSDESEEPQKKKAVKNSKESSDSSEEDSEDESEEKPSKTPQKRGRDV 755
Query: 1059 GMVDA 1063
MVDA
Sbjct: 756 EMVDA 770
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,190,074
Number of Sequences: 63676
Number of extensions: 759011
Number of successful extensions: 3833
Number of sequences better than 10.0: 124
Number of HSP's better than 10.0 without gapping: 3437
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3682
length of query: 1389
length of database: 12,639,632
effective HSP length: 109
effective length of query: 1280
effective length of database: 5,698,948
effective search space: 7294653440
effective search space used: 7294653440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0028b.5


