

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.4
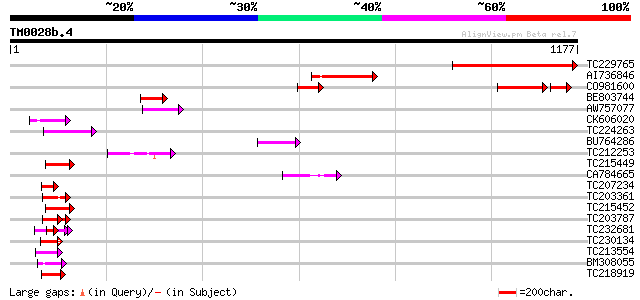
(1177 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229765 similar to UP|Q7NHE2 (Q7NHE2) Gll2595 protein, partial ... 436 e-122
AI736846 224 1e-58
CO981600 165 1e-53
BE803744 112 1e-24
AW757077 70 4e-12
CK606020 55 2e-07
TC224263 weakly similar to UP|Q6GQH4 (Q6GQH4) Egr1 protein, part... 52 2e-06
BU764286 51 3e-06
TC212253 homologue to UP|Q9FNQ1 (Q9FNQ1) RNA helicase, partial (... 49 2e-05
TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%) 47 5e-05
CA784665 47 6e-05
TC207234 weakly similar to UP|Q9FH01 (Q9FH01) Similarity to CHP-... 47 6e-05
TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 46 8e-05
TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%) 46 1e-04
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 45 1e-04
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 45 2e-04
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 45 2e-04
TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protei... 45 2e-04
BM308055 44 3e-04
TC218919 homologue to UP|Q39892 (Q39892) Nucleosome assembly pro... 44 3e-04
>TC229765 similar to UP|Q7NHE2 (Q7NHE2) Gll2595 protein, partial (5%)
Length = 1012
Score = 436 bits (1122), Expect = e-122
Identities = 220/259 (84%), Positives = 235/259 (89%)
Frame = +1
Query: 919 RDAIEQYKDQRNKVSRLKKRISRTEGYKEYNKIIDGVKFIEEKIKRLKTRSKRLTNRIEQ 978
+DAIE+YK+QRNKVSRLKK+I R+EGYKEY KIID VKF EEKIKRLK RSKRL NRIEQ
Sbjct: 1 KDAIERYKEQRNKVSRLKKKIVRSEGYKEYFKIIDAVKFTEEKIKRLKNRSKRLINRIEQ 180
Query: 979 IEPSGWKEFMQVSNVIHETRALDINTHVIFPLGETAAAIRGENELWLAMVLRSKILVGLK 1038
IEPSGWKEFMQVSNVIHE RA DINTH+IFPLGETAAAIRGENELWLAMVLR+KIL+ LK
Sbjct: 181 IEPSGWKEFMQVSNVIHEIRA*DINTHIIFPLGETAAAIRGENELWLAMVLRNKILLELK 360
Query: 1039 PPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSATVVNCIGLLGEQRSALLAIQEKHGVTIS 1098
P QLAAVCA LVS GIKVRP KNNS+IYEPSATV I LL EQRSALLA+Q+KH VTIS
Sbjct: 361 PAQLAAVCASLVSAGIKVRPGKNNSYIYEPSATVTKFITLLDEQRSALLAMQDKHEVTIS 540
Query: 1099 CCLDTQFCGMVEAWASGLTWREIMMDCAMDDGDLARLLRRTIDLLAQIPKLADIDPLLQR 1158
CCLD+QFCGMVEAWASGLTWRE+MMDCAMDDGDLARLLRRTIDLL QIPKL DIDPLL+
Sbjct: 541 CCLDSQFCGMVEAWASGLTWRELMMDCAMDDGDLARLLRRTIDLLVQIPKLPDIDPLLKH 720
Query: 1159 NARAASDVMDRPPISELAG 1177
NA+AAS VMDRPPISEL G
Sbjct: 721 NAKAASSVMDRPPISELVG 777
>AI736846
Length = 409
Score = 224 bits (572), Expect = 1e-58
Identities = 117/137 (85%), Positives = 128/137 (93%)
Frame = +1
Query: 626 SGVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKKNEKEIEL 685
+GVKAIHRSNESD+M+ STG +TLE+ARKLVEQSFGNYVSSNVMLAAKEE+ K EKEIE
Sbjct: 1 AGVKAIHRSNESDNMKPSTG-KTLEEARKLVEQSFGNYVSSNVMLAAKEEINKIEKEIEF 177
Query: 686 LMSEITDEAIDRKSRKALSQKQYKEIAELQEDLRAEKRVRTALRRQMEAKRISALKPLLD 745
LMSEITDEAIDRKSRKALS +QYKEIAEL EDLRAEKRVR+ LR+Q EAKRISALKPLL+
Sbjct: 178 LMSEITDEAIDRKSRKALSPRQYKEIAELLEDLRAEKRVRSELRKQKEAKRISALKPLLE 357
Query: 746 DPESGHLPFLCLQYRDS 762
+PESGHLPFLCLQYRDS
Sbjct: 358 EPESGHLPFLCLQYRDS 408
>CO981600
Length = 852
Score = 165 bits (418), Expect(2) = 1e-53
Identities = 84/105 (80%), Positives = 92/105 (87%)
Frame = -2
Query: 1012 ETAAAIRGENELWLAMVLRSKILVGLKPPQLAAVCAGLVSEGIKVRPWKNNSFIYEPSAT 1071
+TAAAIRGENELWLAMVL SKIL+ LKP QLAAVCA LVS GIKVRP KNNS+IY+PSAT
Sbjct: 698 KTAAAIRGENELWLAMVLCSKILLELKPAQLAAVCASLVSAGIKVRPGKNNSYIYKPSAT 519
Query: 1072 VVNCIGLLGEQRSALLAIQEKHGVTISCCLDTQFCGMVEAWASGL 1116
V I LL EQR+ALLA+Q+KH VTISCCLD+QFCG VEAWASGL
Sbjct: 518 VTKFITLLDEQRNALLAMQDKHEVTISCCLDSQFCGKVEAWASGL 384
Score = 91.7 bits (226), Expect = 2e-18
Identities = 47/54 (87%), Positives = 51/54 (94%)
Frame = -1
Query: 598 ECCKVLFAGLEPLVSQFTASYGMVLNLLSGVKAIHRSNESDDMRQSTGGRTLED 651
E CKVLFAGLEPLVSQFTASYGMVLNLL+GVKAIHRSNESD+M+ ST G+TLED
Sbjct: 852 EGCKVLFAGLEPLVSQFTASYGMVLNLLAGVKAIHRSNESDNMKPST-GKTLED 694
Score = 64.3 bits (155), Expect(2) = 1e-53
Identities = 31/45 (68%), Positives = 35/45 (76%)
Frame = -3
Query: 1122 MMDCAMDDGDLARLLRRTIDLLAQIPKLADIDPLLQRNARAASDV 1166
++ CAMDDGDLARLLRRTIDLL QIPKL DIDPLL + + V
Sbjct: 394 LLGCAMDDGDLARLLRRTIDLLVQIPKLPDIDPLLNLSGNLFASV 260
>BE803744
Length = 275
Score = 112 bits (279), Expect = 1e-24
Identities = 53/56 (94%), Positives = 54/56 (95%)
Frame = +2
Query: 272 VNVDVIVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQI 327
VNVDVIVLDEVHYLSDIS GTVWEEIVIYCPKEV+LICLSATVANPDELAGWIG I
Sbjct: 17 VNVDVIVLDEVHYLSDISHGTVWEEIVIYCPKEVKLICLSATVANPDELAGWIGHI 184
>AW757077
Length = 423
Score = 70.5 bits (171), Expect = 4e-12
Identities = 37/88 (42%), Positives = 52/88 (59%), Gaps = 3/88 (3%)
Frame = +1
Query: 277 IVLDEVHYLSDISRGTVWEEIVIYCPKEVQLICLSATVANPDELAGWIGQIHGK-TELVT 335
IV DEVHY+ D RG VWEE ++ PK + + LSATV N E A W+ ++H + +V
Sbjct: 4 IVFDEVHYMRDRERGVVWEESIVLSPKNSRFVFLSATVPNAKEFADWVAKVHQQPCHVVY 183
Query: 336 SSKRPVPLTWHF--SMKNSLLPLLDEKG 361
+ RP PL + S + L ++DEKG
Sbjct: 184 TDYRPTPLQHYLFPSGGDGLYLVVDEKG 267
>CK606020
Length = 340
Score = 54.7 bits (130), Expect = 2e-07
Identities = 30/85 (35%), Positives = 51/85 (59%)
Frame = +1
Query: 41 LRSLSSTLKFRLSFSFNPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
L S S +L+F++ F NP SLP A +D+++ D++EEEEE EEEE E++++++
Sbjct: 94 LTSFSHSLRFQIQF--NP---MSLPDSPVANDDEIDPSLDEEEEEEEEEEEEEEEEEEEE 258
Query: 101 VAADEYDDEVPDEGEAFDASARREE 125
+ +E ++E E E + EE
Sbjct: 259 IEEEEEEEEEEVEEEVEEEEVEGEE 333
>TC224263 weakly similar to UP|Q6GQH4 (Q6GQH4) Egr1 protein, partial (8%)
Length = 790
Score = 51.6 bits (122), Expect = 2e-06
Identities = 32/112 (28%), Positives = 63/112 (55%), Gaps = 3/112 (2%)
Frame = -2
Query: 71 GEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQR 130
G++D + D+D+ E++E E+E+ +DD D+D ++EY++ + + +A E
Sbjct: 642 GKEDEQGSDNDENEDDEDEDEDSDDDYDEDSGSEEYEETEEEFLNRYAKAAEALENGSAI 463
Query: 131 VEKLCNEVREFGAE---IIDVDELASVYDFRIDKFQRLAVQAFLRGSSVVVS 179
+E+ +E +E G E +IDVDE +V IDK+ + + + S +V++
Sbjct: 462 IEEGDDEDQELGLELGQLIDVDE-QNVLLSLIDKYHHVLTRGLVLPSELVMN 310
>BU764286
Length = 428
Score = 51.2 bits (121), Expect = 3e-06
Identities = 28/91 (30%), Positives = 47/91 (50%), Gaps = 3/91 (3%)
Frame = +1
Query: 515 KAFIEELFQKGLVKVVFATETLAAGINMPARTAVISSLSKRIDSGRR--PLSSNELLQMA 572
++ +EELF ++++ T TLA G+N+PA +I +R ++LQM
Sbjct: 13 RSLVEELFANNKIQILVCTSTLAWGVNLPAHLVIIKGTEYYDGKAKRYVDFPITDILQMM 192
Query: 573 GRAGRRGIDESGH-VVLIQTPNEGAEECCKV 602
GRAGR D+ G V+L+ P + + K+
Sbjct: 193 GRAGRPQFDQHGKAVILVHEPKKSFYKKVKI 285
>TC212253 homologue to UP|Q9FNQ1 (Q9FNQ1) RNA helicase, partial (11%)
Length = 703
Score = 48.5 bits (114), Expect = 2e-05
Identities = 43/155 (27%), Positives = 68/155 (43%), Gaps = 14/155 (9%)
Frame = +1
Query: 203 RIFYTTPLKALSNQKFREFRETF----GDSYV---GLLTGDSAVNKDAQVLIMTTEILRN 255
++ Y PLKA+ ++ ++++ G V G T D A ++I T E
Sbjct: 46 KVIYIAPLKAIVRERMSDWQKRLVSQLGKKMVEMTGDYTPDLTALLSANIIISTPE---- 213
Query: 256 MLYQSVGNVSSSRSGLVNVDVIVLDEVHYLSDISRGTVWEEIV-------IYCPKEVQLI 308
+ + SRS + V +++LDE+H L RG + E IV + V+ +
Sbjct: 214 -KWDGISRNWHSRSYVTKVGLMILDEIHLLG-ADRGPILEVIVSRMRYISSQTERAVRFV 387
Query: 309 CLSATVANPDELAGWIGQIHGKTELVTSSKRPVPL 343
LS +AN +LA W+G S RPVPL
Sbjct: 388 GLSTALANAGDLADWLGVEEIGLFNFKPSVRPVPL 492
>TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%)
Length = 1495
Score = 47.0 bits (110), Expect = 5e-05
Identities = 23/60 (38%), Positives = 37/60 (61%)
Frame = +2
Query: 74 DVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEK 133
D+ED++DD++ EE+ +EEE ED+DDDD DE + + + A S R + Q+ E+
Sbjct: 1037 DLEDDEDDEDIEEDDDEEEEEDEDDDDDEDDEEESKTKKKSSAPKKSGRAQLGDGQQGER 1216
Score = 42.0 bits (97), Expect = 0.002
Identities = 16/29 (55%), Positives = 25/29 (86%)
Frame = +2
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
EDD +DED +++++EE EE+E +DDD+DD
Sbjct: 1043 EDDEDDEDIEEDDDEEEEEDEDDDDDEDD 1129
Score = 41.6 bits (96), Expect = 0.002
Identities = 25/74 (33%), Positives = 39/74 (51%), Gaps = 4/74 (5%)
Frame = +2
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEA----FDASARREEFK 127
EDD + E+DDDEEEEE E+++ ++DD+++ + G A R E K
Sbjct: 1052 EDDEDIEEDDDEEEEEDEDDDDDEDDEEESKTKKKSSAPKKSGRAQLGDGQQGERPPECK 1231
Query: 128 WQRVEKLCNEVREF 141
Q ++K+ V EF
Sbjct: 1232 QQ*MQKVIYVVVEF 1273
Score = 32.7 bits (73), Expect = 0.96
Identities = 16/43 (37%), Positives = 27/43 (62%)
Frame = +2
Query: 71 GEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDE 113
GE DE D E++E+ E+ E +DD++++ D+ DDE +E
Sbjct: 1007 GEAIQGDEFGDLEDDEDDEDIEEDDDEEEEEDEDDDDDEDDEE 1135
>CA784665
Length = 434
Score = 46.6 bits (109), Expect = 6e-05
Identities = 35/123 (28%), Positives = 54/123 (43%)
Frame = +1
Query: 567 ELLQMAGRAGRRGIDESGHVVLIQTPNEGAEECCKVLFAGLEPLVSQFTASYGMVLNLLS 626
E +QM+GRAGRRGIDE G +L+ ++ + L S F SY M+LN
Sbjct: 4 EYIQMSGRAGRRGIDERGICILMVDEKMEPSTAKNMVKGAADSLNSAFHLSYNMILN--- 174
Query: 627 GVKAIHRSNESDDMRQSTGGRTLEDARKLVEQSFGNYVSSNVMLAAKEELKKNEKEIELL 686
MR G D L+ SF + + + ++++K E+E E +
Sbjct: 175 ------------QMRCEDG-----DPENLLRNSFFQFQADRAIPDLEKQIKSLEEERESI 303
Query: 687 MSE 689
+ E
Sbjct: 304 VIE 312
>TC207234 weakly similar to UP|Q9FH01 (Q9FH01) Similarity to CHP-rich zinc
finger protein, partial (4%)
Length = 449
Score = 46.6 bits (109), Expect = 6e-05
Identities = 20/34 (58%), Positives = 25/34 (72%)
Frame = +1
Query: 67 FSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
FS +DD DEDDD +E+ + EEE+Y DDDDDD
Sbjct: 175 FSSEKKDDHSDEDDDGDEDYDDEEEDYVDDDDDD 276
>TC203361 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 1090
Score = 46.2 bits (108), Expect = 8e-05
Identities = 23/57 (40%), Positives = 36/57 (62%)
Frame = -1
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREE 125
D +DD +D+DDDD+EE+E EEE ED +D+ + +DE +E EA +R++
Sbjct: 394 DDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDE---ENEEDEEDEEEEALQPPKKRKK 233
Score = 42.4 bits (98), Expect = 0.001
Identities = 19/64 (29%), Positives = 39/64 (60%)
Frame = -1
Query: 70 AGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQ 129
A E+ EDED+D +++++ E+++ +DDD++D +E D++ ++ E + EE Q
Sbjct: 433 AEENGEEDEDEDGDDQDDDEDDDDDDDDEEDEGGEEEDEDGAEDEENEEDEEDEEEEALQ 254
Query: 130 RVEK 133
+K
Sbjct: 253 PPKK 242
Score = 40.0 bits (92), Expect = 0.006
Identities = 21/69 (30%), Positives = 34/69 (48%)
Frame = -1
Query: 57 NPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEA 116
N S+ + G E+ ++DE+E+ ++++ EDDDDDD DDE + GE
Sbjct: 487 NSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDEDDDDDD------DDEEDEGGEE 326
Query: 117 FDASARREE 125
D +E
Sbjct: 325 EDEDGAEDE 299
Score = 40.0 bits (92), Expect = 0.006
Identities = 20/71 (28%), Positives = 39/71 (54%)
Frame = -1
Query: 55 SFNPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEG 114
S N S P G ++ +ED+D++ +++ ++E+ +DDDDD+ DE +E ++G
Sbjct: 484 SNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDEDDDDDDDDE--EDEGGEEEDEDG 311
Query: 115 EAFDASARREE 125
+ + EE
Sbjct: 310 AEDEENEEDEE 278
>TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%)
Length = 609
Score = 45.8 bits (107), Expect = 1e-04
Identities = 22/60 (36%), Positives = 37/60 (61%)
Frame = +3
Query: 74 DVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEK 133
D+ED++DD++ EE+ +E+E +DDDDDD DE + + + A S R + Q+ E+
Sbjct: 132 DLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEEESKTKKKSSAPKKSGRAQLGDGQQGER 311
Score = 40.0 bits (92), Expect = 0.006
Identities = 14/29 (48%), Positives = 25/29 (85%)
Frame = +3
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
EDD +DED +++++E+ E+++ +DDDDDD
Sbjct: 138 EDDEDDEDIEEDDDEDEEDDDDDDDDDDD 224
Score = 38.9 bits (89), Expect = 0.013
Identities = 19/46 (41%), Positives = 31/46 (67%)
Frame = +3
Query: 71 GEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEA 116
G++ + EDD+D+E+ E +++E E+DDDDD DD+ DE E+
Sbjct: 117 GDEFGDLEDDEDDEDIEEDDDEDEEDDDDD------DDDDDDEEES 236
Score = 38.9 bits (89), Expect = 0.013
Identities = 17/41 (41%), Positives = 29/41 (70%)
Frame = +3
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDE 109
+A + D + +DDE++E+ EE++ ED++DDD D+ DDE
Sbjct: 105 EAIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDE 227
Score = 32.3 bits (72), Expect = 1.3
Identities = 16/43 (37%), Positives = 27/43 (62%)
Frame = +3
Query: 71 GEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDE 113
GE DE D E++E+ E+ E +DD+D++ D+ DD+ +E
Sbjct: 102 GEAIQGDEFGDLEDDEDDEDIEEDDDEDEEDDDDDDDDDDDEE 230
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 45.4 bits (106), Expect = 1e-04
Identities = 20/41 (48%), Positives = 29/41 (69%)
Frame = -3
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDE 109
D +DD ED+DDDDEE+E EEE+ + +D++ DE D+E
Sbjct: 427 DDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEE 305
Score = 45.1 bits (105), Expect = 2e-04
Identities = 18/57 (31%), Positives = 38/57 (66%)
Frame = -3
Query: 69 DAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREE 125
D ED + +DDD++++++ EE+E +++D+D A DE ++E ++ EA +R++
Sbjct: 445 DEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAEDEENEEDEEDEEALQPPKKRKK 275
Score = 42.4 bits (98), Expect = 0.001
Identities = 21/71 (29%), Positives = 38/71 (52%)
Frame = -3
Query: 55 SFNPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEG 114
S N S P G ++ +ED+D++ +++ +++E +DDDD++ A E +DE E
Sbjct: 517 SNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAED 338
Query: 115 EAFDASARREE 125
E + EE
Sbjct: 337 EENEEDEEDEE 305
Score = 39.7 bits (91), Expect = 0.008
Identities = 18/69 (26%), Positives = 36/69 (52%)
Frame = -3
Query: 57 NPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEA 116
N S+ + G E+ ++DE+E+ ++++ ++DDDDD DE +E ++G
Sbjct: 520 NSNNKSNSKKAPEGGAGGAEENGEEDEDEDGDDQDDDDEDDDDDDEEDEAGEEEDEDGAE 341
Query: 117 FDASARREE 125
+ + EE
Sbjct: 340 DEENEEDEE 314
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 45.1 bits (105), Expect = 2e-04
Identities = 26/74 (35%), Positives = 36/74 (48%)
Frame = +1
Query: 51 RLSFSFNPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEV 110
RLSF + D D +D+DD+DEEEE + E DDDD+ DE +DE
Sbjct: 199 RLSFDDDDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQRGGEPDDDDNDDDDEDEDEE 378
Query: 111 PDEGEAFDASARRE 124
+E + +A E
Sbjct: 379 DEEEQGEEADLGTE 420
Score = 35.0 bits (79), Expect = 0.19
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Frame = +1
Query: 71 GEDDVEDEDDDDEEEEEY--EEEEYEDDDDDDVAADEYDDEVPDE 113
GE++ ED DD+D E+ E + + + DD DD E DDE P +
Sbjct: 490 GEEEEEDVDDEDGEKSEAPPKRKRSDKDDSDDDDGGEDDDERPSK 624
Score = 32.7 bits (73), Expect = 0.96
Identities = 15/27 (55%), Positives = 20/27 (73%), Gaps = 1/27 (3%)
Frame = +2
Query: 81 DDEEEEEYE-EEEYEDDDDDDVAADEY 106
D+EE+EEYE E+E DD +DD D+Y
Sbjct: 125 DEEEDEEYESEDEVVDDGEDDEDDDDY 205
Score = 32.3 bits (72), Expect(2) = 5e-04
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +1
Query: 93 YEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQR 130
++DDDD++ D+ +D+ PD G+ D EE QR
Sbjct: 208 FDDDDDNEDEEDDDEDDAPDGGDDDDDEDEEEESDVQR 321
Score = 31.6 bits (70), Expect = 2.1
Identities = 28/99 (28%), Positives = 34/99 (34%), Gaps = 39/99 (39%)
Frame = +1
Query: 69 DAGEDDVEDEDDDDEEE------------------------------EEYEEEEYEDDDD 98
D +DD EDED++DEEE EE EEE ED DD
Sbjct: 340 DDNDDDDEDEDEEDEEEQGEEADLGTEYLIRPLVTAEEEEASSDFEPEENGEEEEEDVDD 519
Query: 99 DDVAADE---------YDDEVPDEGEAFDASARREEFKW 128
+D E DD D+G D + W
Sbjct: 520 EDGEKSEAPPKRKRSDKDDSDDDDGGEDDDERPSKR*NW 636
Score = 30.8 bits (68), Expect = 3.7
Identities = 12/39 (30%), Positives = 23/39 (58%)
Frame = +1
Query: 68 SDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEY 106
SD D+DD+D+++E+ +EE+ E+ ++ EY
Sbjct: 307 SDVQRGGEPDDDDNDDDDEDEDEEDEEEQGEEADLGTEY 423
Score = 30.4 bits (67), Expect(2) = 5e-04
Identities = 11/25 (44%), Positives = 19/25 (76%)
Frame = +2
Query: 76 EDEDDDDEEEEEYEEEEYEDDDDDD 100
E+ED++ E E+E ++ +D+DDDD
Sbjct: 128 EEEDEEYESEDEVVDDGEDDEDDDD 202
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 44.7 bits (104), Expect = 2e-04
Identities = 20/45 (44%), Positives = 31/45 (68%)
Frame = +2
Query: 65 PTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDE 109
P + GE D +DEDDDD++++ E++ EDDDD++ DE +DE
Sbjct: 398 PEANGGGESDDDDEDDDDDDDDNDEDDGDEDDDDEE---DEDEDE 523
Score = 38.9 bits (89), Expect = 0.013
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 25/75 (33%)
Frame = +2
Query: 66 TFSDAGEDDV---EDEDDDDEEEEEYE----------------------EEEYEDDDDDD 100
T D +DDV ED DDDDE++E++ +++ EDDDDDD
Sbjct: 278 TEDDDDDDDVNDGEDGDDDDEDDEDFSGDDGGEEADSDDDPEANGGGESDDDDEDDDDDD 457
Query: 101 VAADEYDDEVPDEGE 115
DE D + D+ E
Sbjct: 458 DDNDEDDGDEDDDDE 502
Score = 38.5 bits (88), Expect = 0.018
Identities = 20/56 (35%), Positives = 30/56 (52%)
Frame = +2
Query: 70 AGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREE 125
+G+D E+ D DD+ E E +DD+DDD D+ DD D+G+ D E+
Sbjct: 356 SGDDGGEEADSDDDPEANGGGESDDDDEDDD---DDDDDNDEDDGDEDDDDEEDED 514
Score = 36.6 bits (83), Expect = 0.067
Identities = 19/64 (29%), Positives = 34/64 (52%), Gaps = 8/64 (12%)
Frame = +2
Query: 69 DAGEDDVEDED--------DDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDAS 120
D GE+ D+D DD++E++ ++++ D+DD D D+ +DE DE S
Sbjct: 365 DGGEEADSDDDPEANGGGESDDDDEDDDDDDDDNDEDDGDEDDDDEEDEDEDEETPQPPS 544
Query: 121 ARRE 124
+R+
Sbjct: 545 KKRK 556
Score = 35.8 bits (81), Expect = 0.11
Identities = 16/62 (25%), Positives = 32/62 (50%)
Frame = +2
Query: 54 FSFNPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDE 113
F + P + + D ED+DDDD+ + E+ + +D+DD+D + D+ +E +
Sbjct: 215 FPLDQPCKGKHTSEENKDASDTEDDDDDDDVNDG-EDGDDDDEDDEDFSGDDGGEEADSD 391
Query: 114 GE 115
+
Sbjct: 392 DD 397
Score = 33.9 bits (76), Expect = 0.43
Identities = 11/44 (25%), Positives = 30/44 (68%)
Frame = +2
Query: 57 NPPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDD 100
+P + + D +DD +D+D+D+++ +E +++E ++D+D++
Sbjct: 395 DPEANGGGESDDDDEDDDDDDDDNDEDDGDEDDDDEEDEDEDEE 526
Score = 33.5 bits (75), Expect = 0.56
Identities = 15/45 (33%), Positives = 28/45 (61%)
Frame = +2
Query: 74 DVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFD 118
D D +DDD++++ + E+ +DDD+DD D D+ +E ++ D
Sbjct: 266 DASDTEDDDDDDDVNDGEDGDDDDEDD--EDFSGDDGGEEADSDD 394
>TC213554 weakly similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1,
partial (18%)
Length = 451
Score = 44.7 bits (104), Expect = 2e-04
Identities = 23/60 (38%), Positives = 36/60 (59%), Gaps = 3/60 (5%)
Frame = +2
Query: 53 SFSFNPPTSSSLPTFSDAG---EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDE 109
S S PP P +A E++VE+E +++E EEE EEEE E++D++D +E D +
Sbjct: 125 SVSAEPPLKQHQPKPENAAQEYEEEVEEEVEEEEVEEEVEEEEEEEEDEEDEEEEEEDQK 304
Score = 38.1 bits (87), Expect = 0.023
Identities = 17/63 (26%), Positives = 38/63 (59%), Gaps = 2/63 (3%)
Frame = +2
Query: 58 PPTSSSLPTFSDAGEDDVEDEDDDDE--EEEEYEEEEYEDDDDDDVAADEYDDEVPDEGE 115
P ++ + + E++VE+E+ ++E EEEE EE+E +++++++ D+Y + + E
Sbjct: 161 PKPENAAQEYEEEVEEEVEEEEVEEEVEEEEEEEEDEEDEEEEEEDQKDQYQHQQQQQDE 340
Query: 116 AFD 118
D
Sbjct: 341 DDD 349
Score = 33.1 bits (74), Expect = 0.74
Identities = 13/38 (34%), Positives = 24/38 (62%)
Frame = +2
Query: 73 DDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEV 110
++ E+E++D+E+EEE EE++ + DE DD +
Sbjct: 242 EEEEEEEEDEEDEEEEEEDQKDQYQHQQQQQDEDDDPI 355
>BM308055
Length = 416
Score = 44.3 bits (103), Expect = 3e-04
Identities = 24/65 (36%), Positives = 38/65 (57%), Gaps = 4/65 (6%)
Frame = +2
Query: 58 PPTSSSLPTFSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDD----DDVAADEYDDEVPDE 113
PP + F D ++ E+EDD D E + ++++E E+DDD DD+ A +Y +E PD+
Sbjct: 68 PPNGDPVRAFVD---EEAEEEDDSDNELQLFQDDEGEEDDDDIEEDDMIATQY-EEKPDD 235
Query: 114 GEAFD 118
E D
Sbjct: 236 REKHD 250
Score = 33.9 bits (76), Expect = 0.43
Identities = 16/55 (29%), Positives = 29/55 (52%)
Frame = +2
Query: 72 EDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEF 126
+DD +EDDDD EE++ +YE+ DD D+ + ++ +A ++F
Sbjct: 152 QDDEGEEDDDDIEEDDMIATQYEEKPDDREKHDQLHQQWLEQQDAAGMDNLHQKF 316
>TC218919 homologue to UP|Q39892 (Q39892) Nucleosome assembly protein 1,
partial (33%)
Length = 729
Score = 44.3 bits (103), Expect = 3e-04
Identities = 19/50 (38%), Positives = 34/50 (68%)
Frame = +1
Query: 67 FSDAGEDDVEDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDEVPDEGEA 116
F D +D+ E+ED+D++E+EEY+E+E +D+++DD + V G+A
Sbjct: 178 FEDLEDDEDEEEDEDEDEDEEYDEDE-DDEEEDDTKTKKKLSAVKKSGKA 324
Score = 39.3 bits (90), Expect = 0.010
Identities = 21/49 (42%), Positives = 31/49 (62%), Gaps = 3/49 (6%)
Frame = +1
Query: 59 PTSSSLPTFSDAGEDDVEDEDDDDEEEE---EYEEEEYEDDDDDDVAAD 104
P + S T A D+ ED +DD++EEE E E+EEY++D+DD+ D
Sbjct: 130 PHAVSWFTGEXAQGDEFEDLEDDEDEEEDEDEDEDEEYDEDEDDEEEDD 276
Score = 34.7 bits (78), Expect = 0.25
Identities = 15/34 (44%), Positives = 25/34 (73%)
Frame = +1
Query: 76 EDEDDDDEEEEEYEEEEYEDDDDDDVAADEYDDE 109
E ED +D+E+EE +E+E ED++ D+ DE +D+
Sbjct: 175 EFEDLEDDEDEEEDEDEDEDEEYDEDEDDEEEDD 276
Score = 32.7 bits (73), Expect = 0.96
Identities = 19/57 (33%), Positives = 31/57 (54%)
Frame = +1
Query: 86 EEYEEEEYEDDDDDDVAADEYDDEVPDEGEAFDASARREEFKWQRVEKLCNEVREFG 142
E + +E+ED +DD+ DE +DE DE E +D EE + +K + V++ G
Sbjct: 157 EXAQGDEFEDLEDDE---DEEEDEDEDEDEEYDEDEDDEEEDDTKTKKKLSAVKKSG 318
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,005,125
Number of Sequences: 63676
Number of extensions: 668140
Number of successful extensions: 9775
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 5153
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7106
length of query: 1177
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1069
effective length of database: 5,762,624
effective search space: 6160245056
effective search space used: 6160245056
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0028b.4


