

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.3
(288 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
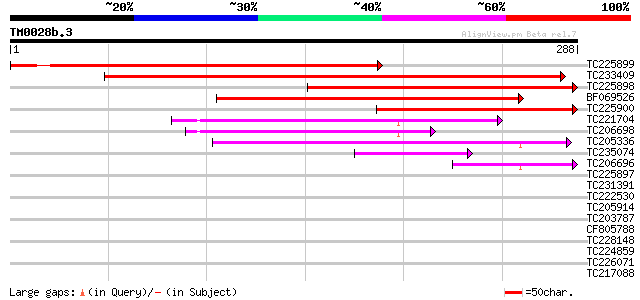
TC225899 similar to UP|O48685 (O48685) Pyruvate dehydrogenase E1... 296 7e-81
TC233409 weakly similar to UP|ODPA_SMIMA (P52900) Pyruvate dehyd... 284 3e-77
TC225898 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase ... 270 7e-73
BF069526 weakly similar to SP|P52902|ODPA Pyruvate dehydrogenase... 199 1e-51
TC225900 homologue to UP|ODPA_PEA (P52902) Pyruvate dehydrogenas... 198 2e-51
TC221704 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase ... 151 3e-37
TC206698 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase ... 119 2e-27
TC205336 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto... 94 9e-20
TC235074 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto... 49 3e-06
TC206696 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1... 48 5e-06
TC225897 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase ... 38 0.005
TC231391 UP|Q39846 (Q39846) LEA protein, complete 32 0.33
TC222530 weakly similar to UP|Q9LVF9 (Q9LVF9) Gb|AAB82628.1, par... 31 0.74
TC205914 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, ... 30 0.97
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 30 0.97
CF805788 30 1.3
TC228148 similar to UP|Q7YXC8 (Q7YXC8) C. elegans GRD-1 protein ... 30 1.7
TC224859 weakly similar to UP|Q7TP79 (Q7TP79) Aa2-245, partial (3%) 30 1.7
TC226071 weakly similar to GB|AAM98298.1|22655412|AY142034 At5g0... 29 2.2
TC217088 UP|Q9FQD5 (Q9FQD5) Glutathione S-transferase GST 23 , ... 28 3.7
>TC225899 similar to UP|O48685 (O48685) Pyruvate dehydrogenase E1 alpha
subunit , partial (40%)
Length = 700
Score = 296 bits (758), Expect = 7e-81
Identities = 147/190 (77%), Positives = 164/190 (85%), Gaps = 1/190 (0%)
Frame = +3
Query: 1 MALSRLASSSSSSSSTVGSKFLKPLTAAFSLRR-SISSDSNPITIETSVPFTAHNCDPPS 59
MALSR+ + SS S+ LKPL + SLRR S+S+ S+P+TIETSVPFT+HNCD PS
Sbjct: 147 MALSRVVAQSSQSN------LLKPLASYLSLRRRSVSTSSDPLTIETSVPFTSHNCDAPS 308
Query: 60 RSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKK 119
R+VETS+AEL +FF MA MRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEA IT+K
Sbjct: 309 RAVETSSAELFAFFHDMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITRK 488
Query: 120 DAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGA 179
D +ITAYRDHCTFL RGGTL+EIFSELMGR DGCSKGKGGSMHFYRKE FYGGHGIVGA
Sbjct: 489 DCVITAYRDHCTFLARGGTLIEIFSELMGRRDGCSKGKGGSMHFYRKEGGFYGGHGIVGA 668
Query: 180 QVPLGCGVAF 189
QVPLGCG+AF
Sbjct: 669 QVPLGCGLAF 698
>TC233409 weakly similar to UP|ODPA_SMIMA (P52900) Pyruvate dehydrogenase E1
component alpha subunit, mitochondrial precursor
(PDHE1-A) (Fragment) , partial (66%)
Length = 1139
Score = 284 bits (727), Expect = 3e-77
Identities = 139/235 (59%), Positives = 172/235 (73%), Gaps = 1/235 (0%)
Frame = +3
Query: 49 PFTAHNCDP-PSRSVETSTAELHSFFRQMATMRRMEIAADSLYKAKLIRGFCHLYDGQEA 107
P+ H D PS VET+ +L S F+ M +R+MEI AD +YKAK I+GF HLY+G+EA
Sbjct: 114 PYDLHRLDKGPSLEVETNKEDLVSAFKTMYEIRQMEIIADKMYKAKKIQGFLHLYNGEEA 293
Query: 108 VAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKE 167
V G+E+GI D +ITAYRDH L RGG ++F+EL G+V G S+GKGGSMH Y KE
Sbjct: 294 VVTGIESGIRPDDHVITAYRDHGFILTRGGNAEQVFAELQGKVTGTSRGKGGSMHMYSKE 473
Query: 168 TNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLP 227
FYGG+GIVGAQ P+G G+AFA KY+K + V YGDG+ANQGQLFEA N+AALW LP
Sbjct: 474 GKFYGGNGIVGAQTPVGAGLAFAAKYNKTDQVCITCYGDGSANQGQLFEAYNMAALWKLP 653
Query: 228 AILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEH 282
I VCENN YGMGT+ RAA S +Y RGD+VPG+++DGMD LAVK A KFA ++
Sbjct: 654 VIFVCENNKYGMGTSIDRAAASTTFYTRGDFVPGIRIDGMDYLAVKNATKFAADY 818
>TC225898 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component
alpha subunit, mitochondrial precursor (PDHE1-A) ,
partial (62%)
Length = 1250
Score = 270 bits (689), Expect = 7e-73
Identities = 125/137 (91%), Positives = 131/137 (95%)
Frame = +1
Query: 152 GCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQ 211
GCSKGKGGSMHFYRKE FYGGHGIVGAQVPLGCG+AFAQKY KDE VTF++YGDGAANQ
Sbjct: 1 GCSKGKGGSMHFYRKEGGFYGGHGIVGAQVPLGCGLAFAQKYCKDENVTFSMYGDGAANQ 180
Query: 212 GQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLA 271
GQLFEALNI+ALWDLP+ILVCENNHYGMGTAEWRAAKSP+YYKRGDYVPGLKVDGMD LA
Sbjct: 181 GQLFEALNISALWDLPSILVCENNHYGMGTAEWRAAKSPSYYKRGDYVPGLKVDGMDALA 360
Query: 272 VKQACKFAKEHALKNGP 288
VKQACKFAKE ALKNGP
Sbjct: 361 VKQACKFAKEFALKNGP 411
>BF069526 weakly similar to SP|P52902|ODPA Pyruvate dehydrogenase E1
component alpha subunit mitochondrial precursor (EC
1.2.4.1) (PDHE1-A)., partial (39%)
Length = 469
Score = 199 bits (506), Expect = 1e-51
Identities = 98/156 (62%), Positives = 112/156 (70%)
Frame = +2
Query: 106 EAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYR 165
EA A M+A +T+KD +I YR HCT L GGTL+ + ELM R CS GK G MH YR
Sbjct: 2 EAFAGCMKANLTRKDCLIITYRYHCTLLTLGGTLIYVLPELMRRHHRCSNGKRGCMHLYR 181
Query: 166 KETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWD 225
E+ GGH I GAQVPL C + FAQKY DE V ++YGD AA GQ FEALNI ALWD
Sbjct: 182 TESGLSGGHEIYGAQVPLRCILTFAQKYCIDENVMLSMYGDRAAYLGQWFEALNIYALWD 361
Query: 226 LPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPG 261
L +I+VC+NNHYGMGTAEWRAA SP+YYKR DYVPG
Sbjct: 362 LASIMVCQNNHYGMGTAEWRAANSPSYYKREDYVPG 469
>TC225900 homologue to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1
component alpha subunit, mitochondrial precursor
(PDHE1-A) , partial (54%)
Length = 1042
Score = 198 bits (504), Expect = 2e-51
Identities = 94/102 (92%), Positives = 97/102 (94%)
Frame = +2
Query: 187 VAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRA 246
V + SKDE+VTFA+YGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRA
Sbjct: 8 VGVCSEKSKDESVTFAMYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRA 187
Query: 247 AKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 288
AKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP
Sbjct: 188 AKSPAYYKRGDYVPGLKVDGMDVLAVKQACKFAKEHALKNGP 313
>TC221704 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (46%)
Length = 616
Score = 151 bits (382), Expect = 3e-37
Identities = 72/175 (41%), Positives = 103/175 (58%), Gaps = 7/175 (4%)
Frame = +2
Query: 83 EIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEI 142
++ A Y+ K+ GF HLY+GQEAV+ G + K+D +++ YRDH L +G +
Sbjct: 95 DMCAQMYYRGKMF-GFVHLYNGQEAVSTGFINFLKKEDCVVSTYRDHVHALSKGVPARAV 271
Query: 143 FSELMGRVDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKD------ 196
SEL G+ GCS+G+GGSMH + KE N GG + +P+ G AF+ KY ++
Sbjct: 272 MSELFGKATGCSRGQGGSMHMFSKEHNLIGGFAFIAEGIPVATGAAFSSKYRREVLKEAD 451
Query: 197 -EAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRAAKSP 250
+ VT A +GDG N GQ +E LN+AALW LP + V ENN + +G + RA P
Sbjct: 452 CDHVTLAFFGDGTCNNGQFYECLNMAALWKLPIVFVVENNLWAIGMSHLRATSDP 616
>TC206698 homologue to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (31%)
Length = 523
Score = 119 bits (298), Expect = 2e-27
Identities = 56/134 (41%), Positives = 81/134 (59%), Gaps = 7/134 (5%)
Frame = +3
Query: 90 YKAKLIRGFCHLYDGQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGR 149
Y+ K+ GF HLY+GQEAV+ G + K+D++++ YRDH L +G E+ SEL G+
Sbjct: 15 YRGKMF-GFVHLYNGQEAVSTGFIKLLKKEDSVVSTYRDHVHALSKGVPSREVMSELFGK 191
Query: 150 VDGCSKGKGGSMHFYRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKD-------EAVTFA 202
GC +G+GGSMH + KE N GG +G +P+ G AF+ KY ++ + VT A
Sbjct: 192 ATGCCRGQGGSMHMFSKEHNLLGGFAFIGEGIPVATGAAFSSKYRREVLKQADCDHVTLA 371
Query: 203 LYGDGAANQGQLFE 216
+GDG N GQ +E
Sbjct: 372 FFGDGTCNNGQFYE 413
>TC205336 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein, partial
(93%)
Length = 1642
Score = 93.6 bits (231), Expect = 9e-20
Identities = 54/184 (29%), Positives = 86/184 (46%), Gaps = 2/184 (1%)
Frame = +1
Query: 104 GQEAVAIGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHF 163
G+EAV I A + D I+ YR+ L RG TL + + G KG+ +H+
Sbjct: 583 GEEAVNIASAAALAPDDIILPQYREPGVLLWRGFTLQQFVHQCFGNTHDFGKGRQMPIHY 762
Query: 164 YRKETNFYGGHGIVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAAL 223
+ N++ + Q+P G A++ K A GDGA ++G A+N AA+
Sbjct: 763 GSNQHNYFTVSSPIATQLPQAVGAAYSLKMDGKSACAVTFCGDGATSEGDFHAAMNFAAV 942
Query: 224 WDLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAKE 281
+ P + +C NN + + T +S +G + ++VDG D LAV A A+E
Sbjct: 943 MEAPVVFICRNNGWAISTPVEDQFRSDGIVVKGKAYGIWSIRVDGNDALAVYSAVHTARE 1122
Query: 282 HALK 285
A+K
Sbjct: 1123IAIK 1134
>TC235074 similar to UP|Q9FY85 (Q9FY85) Branched-chain alpha keto-acid
dehydrogenase E1 alpha subunit-like protein, partial
(31%)
Length = 519
Score = 48.5 bits (114), Expect = 3e-06
Identities = 20/60 (33%), Positives = 32/60 (53%)
Frame = +2
Query: 176 IVGAQVPLGCGVAFAQKYSKDEAVTFALYGDGAANQGQLFEALNIAALWDLPAILVCENN 235
+ Q+ G A++ K K +A +GDG +++G ALN AA+ + P I +C NN
Sbjct: 329 VYSTQISHAVGAAYSLKMDKKDACAVTYFGDGGSSEGDFHAALNFAAVLEAPVIFICRNN 508
Score = 33.1 bits (74), Expect = 0.15
Identities = 16/62 (25%), Positives = 28/62 (44%)
Frame = +3
Query: 110 IGMEAGITKKDAIITAYRDHCTFLGRGGTLLEIFSELMGRVDGCSKGKGGSMHFYRKETN 169
+ A + D + YR+ L RG TL E ++L + KG+ H+ K+ N
Sbjct: 3 VASAAALAMDDVVFPQYREAGVLLWRGFTLQEFANQLFSNIYDYGKGRQMPAHYGSKKHN 182
Query: 170 FY 171
++
Sbjct: 183 YF 188
>TC206696 similar to UP|O24457 (O24457) Pyruvate dehydrogenase E1 alpha
subunit (At1g01090/T25K16_8) , partial (43%)
Length = 765
Score = 48.1 bits (113), Expect = 5e-06
Identities = 27/66 (40%), Positives = 38/66 (56%), Gaps = 3/66 (4%)
Frame = +3
Query: 226 LPAILVCENNHYGMGTAEWRAAKSPAYYKRGDY--VPGLKVDGMDVLAVKQACKFAKEHA 283
LP + V ENN + +G + RA P +K+G +PG+ VDGMDVL V++ K A A
Sbjct: 9 LPIVFVVENNLWAIGMSHLRATSDPQIWKKGPAFGMPGVHVDGMDVLQVREVAKEAVGRA 188
Query: 284 LK-NGP 288
+ GP
Sbjct: 189 RRVEGP 206
>TC225897 similar to UP|ODPA_PEA (P52902) Pyruvate dehydrogenase E1 component
alpha subunit, mitochondrial precursor (PDHE1-A) ,
partial (32%)
Length = 820
Score = 38.1 bits (87), Expect = 0.005
Identities = 17/18 (94%), Positives = 17/18 (94%)
Frame = +1
Query: 271 AVKQACKFAKEHALKNGP 288
AVKQACKFAKE ALKNGP
Sbjct: 1 AVKQACKFAKEFALKNGP 54
>TC231391 UP|Q39846 (Q39846) LEA protein, complete
Length = 2208
Score = 32.0 bits (71), Expect = 0.33
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 5/74 (6%)
Frame = -2
Query: 6 LASSSSSSSSTVGSKFLKPLT-----AAFSLRRSISSDSNPITIETSVPFTAHNCDPPSR 60
L+ S S SSS +G P + A L S S+ + P+ + S+PF+A C P S
Sbjct: 716 LSLSLSLSSSLIGF*VPPPFSNVQVITAVVLAPSFSAPTLPLAVSVSLPFSATTCPPLSL 537
Query: 61 SVETSTAELHSFFR 74
+ S + S R
Sbjct: 536 PLSFSVLPITSLVR 495
>TC222530 weakly similar to UP|Q9LVF9 (Q9LVF9) Gb|AAB82628.1, partial (4%)
Length = 639
Score = 30.8 bits (68), Expect = 0.74
Identities = 26/64 (40%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Frame = -3
Query: 7 ASSSSSSSSTVGSKF--LKPLTAAFSLRRSISSDSNPITIETSVP-FTAHNCDPPSRSVE 63
ASS+ +SSST+ S + L P+T+ S R S SS NP T + V T + PSR
Sbjct: 259 ASSARASSSTLASSYSPLHPITS--SPRNSSSSTPNPTTRTSRVRCSTRPPTETPSRGTP 86
Query: 64 TSTA 67
S+A
Sbjct: 85 CSSA 74
>TC205914 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, partial (9%)
Length = 1513
Score = 30.4 bits (67), Expect = 0.97
Identities = 17/34 (50%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Frame = -3
Query: 172 GGH--GIVGAQVPLGCGVAFAQKYSKDEAVTFAL 203
GGH +VG + PLGCGV Y DEAV L
Sbjct: 173 GGHLGAVVGLENPLGCGVG----YELDEAVALGL 84
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 30.4 bits (67), Expect = 0.97
Identities = 23/61 (37%), Positives = 32/61 (51%), Gaps = 3/61 (4%)
Frame = +3
Query: 2 ALSRLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSD---SNPITIETSVPFTAHNCDPP 58
A S +SSS SSSS S P +++ S S SS S+P + +S PF++ PP
Sbjct: 303 ASSSSSSSSFSSSSAPSSSSSSPASSSSSSSSSSSSSSS*SSPSSSSSSSPFSSAPPAPP 482
Query: 59 S 59
S
Sbjct: 483 S 485
>CF805788
Length = 551
Score = 30.0 bits (66), Expect = 1.3
Identities = 18/50 (36%), Positives = 25/50 (50%)
Frame = -3
Query: 5 RLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHN 54
+L S+S + ST+ FLK LTA S D + + S+PFT N
Sbjct: 228 QLLSNSLNLPSTIAGTFLKGLTARNSGDNC*RLDKSTLISSKSIPFTLQN 79
>TC228148 similar to UP|Q7YXC8 (Q7YXC8) C. elegans GRD-1 protein
(Corresponding sequence R08B4.1b), partial (3%)
Length = 1180
Score = 29.6 bits (65), Expect = 1.7
Identities = 23/66 (34%), Positives = 34/66 (50%), Gaps = 5/66 (7%)
Frame = +3
Query: 8 SSSSSSSSTVGSKFLKPLTAAFSLRRS---ISSDSNPITIETSVPFTAHNCDPPSRSV-- 62
SSS + + KP+T A SLRRS +++++N S P T + + PS +V
Sbjct: 210 SSSDTDEEELPKAKEKPVTGARSLRRSYKNLNNNNNKKPSSNSTPKTGSSGNKPSETVKP 389
Query: 63 ETSTAE 68
E S AE
Sbjct: 390 EKSKAE 407
>TC224859 weakly similar to UP|Q7TP79 (Q7TP79) Aa2-245, partial (3%)
Length = 843
Score = 29.6 bits (65), Expect = 1.7
Identities = 16/42 (38%), Positives = 25/42 (59%)
Frame = -1
Query: 5 RLASSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIET 46
R +SSSSSSS T+ S F P + AF + S ++ + ++T
Sbjct: 744 RSSSSSSSSSFTLSSSFSSPSSFAFFCSFPLRSATSSLKVQT 619
>TC226071 weakly similar to GB|AAM98298.1|22655412|AY142034
At5g01750/T20L15_20 {Arabidopsis thaliana;} , partial
(81%)
Length = 1079
Score = 29.3 bits (64), Expect = 2.2
Identities = 15/38 (39%), Positives = 24/38 (62%)
Frame = -2
Query: 27 AAFSLRRSISSDSNPITIETSVPFTAHNCDPPSRSVET 64
A+ +RRS S+P+T++T++P T+ PS SV T
Sbjct: 442 ASTRMRRSRRVMSDPLTLKTTLPLTSVTTKLPSESVIT 329
>TC217088 UP|Q9FQD5 (Q9FQD5) Glutathione S-transferase GST 23 , complete
Length = 1319
Score = 28.5 bits (62), Expect = 3.7
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = -3
Query: 8 SSSSSSSSTVGSKFLKPLTAAFSLRRSISSDSNPITIETSVPFTAHN 54
S+S + ST+ FLK LTA S D + + S+PFT N
Sbjct: 303 SNSLNLPSTIAGTFLKGLTARNSGDNC*RLDKSTLISSKSIPFTLQN 163
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,585,783
Number of Sequences: 63676
Number of extensions: 170770
Number of successful extensions: 1206
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 1057
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1153
length of query: 288
length of database: 12,639,632
effective HSP length: 96
effective length of query: 192
effective length of database: 6,526,736
effective search space: 1253133312
effective search space used: 1253133312
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0028b.3


