

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0028b.2
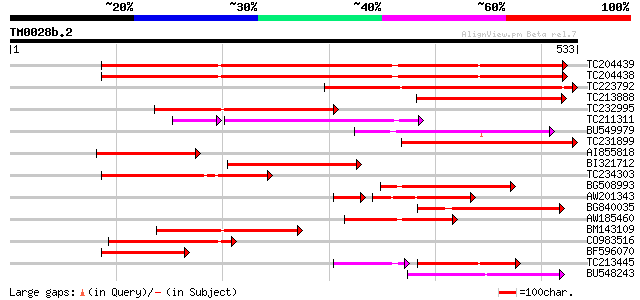
(533 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 374 e-104
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 372 e-103
TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polypro... 250 1e-66
TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%) 159 4e-39
TC232995 154 7e-38
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 129 8e-34
BU549979 132 5e-31
TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Frag... 129 3e-30
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 127 1e-29
BI321712 126 3e-29
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 125 5e-29
BG508993 122 4e-28
AW201343 99 4e-25
BG840035 weakly similar to GP|9294121|dbj copia-like retrotransp... 108 5e-24
AW185460 108 5e-24
BM143109 107 1e-23
CO983516 107 2e-23
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 107 2e-23
TC213445 77 3e-23
BU548243 100 1e-21
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 374 bits (959), Expect = e-104
Identities = 197/439 (44%), Positives = 288/439 (64%), Gaps = 1/439 (0%)
Frame = +1
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G+D+ E FAPVAR+E+IRL++ +A + ++Q+DVKSAFLNG L EEVYV QP GF
Sbjct: 3424 GVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADP 3603
Query: 147 GKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLIC 206
+ V++L KALYGLKQAPRAW +R+ FL Q + + ++ ++V++ L++
Sbjct: 3604 THPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQD--AENLMIAQ 3777
Query: 207 LYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKD 266
+YVDD++ G + L Q ++SEFEMS +G+L YFLG+Q + IF+ Q +Y K+
Sbjct: 3778 IYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKN 3957
Query: 267 ILKRFNMGNCHPVTTPSEVNIKLRRDDEG-DVDASLYKQLVGSLRFLCNSRLDISYGVGL 325
I+K+F M N TP+ ++KL +D+ G VD SLY+ ++GSL +L SR DI+Y VG+
Sbjct: 3958 IVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGV 4137
Query: 326 ISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDR 385
+++ PK SHL KRIL+Y+ GT+DYGI++ LVGY D+D++G DDR
Sbjct: 4138 CARYQANPKISHLTQVKRILKYVNGTSDYGIMYCH----CSNPMLVGYCDADWAGSADDR 4305
Query: 386 KSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKK 445
KSTSG F L ISW SKKQ ++LS+ EAEYIA + Q VW++++LKE V +
Sbjct: 4306 KSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYN-VEQD 4482
Query: 446 PMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTK 505
M L DN+SAI+++KNP+ H R+KHI++R H++R+ V+ + L + T+EQ ADIFTK
Sbjct: 4483 VMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTK 4662
Query: 506 PLKTERFVILRKEIGVVSL 524
L +F LR ++G+ L
Sbjct: 4663 ALDANQFEKLRGKLGICLL 4719
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 372 bits (956), Expect = e-103
Identities = 197/439 (44%), Positives = 288/439 (64%), Gaps = 1/439 (0%)
Frame = +1
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G+D+ E FAPVAR+E+IRL++ +A + ++Q+DVKSAFLNG L EE YV QP GF
Sbjct: 3427 GVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDP 3606
Query: 147 GKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLIC 206
+ V++L KALYGLKQAPRAW +R+ FL Q + + ++ ++V++ L++
Sbjct: 3607 THPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQD--AENLMIAQ 3780
Query: 207 LYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYIKD 266
+YVDD++ G + L Q ++SEFEMS +G+L YFLG+Q + IF+ Q KY K+
Sbjct: 3781 IYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKN 3960
Query: 267 ILKRFNMGNCHPVTTPSEVNIKLRRDDEG-DVDASLYKQLVGSLRFLCNSRLDISYGVGL 325
I+K+F M N TP+ ++KL +D+ G VD SLY+ ++GSL +L SR DI+Y VG+
Sbjct: 3961 IVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGV 4140
Query: 326 ISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDR 385
+++ PK SHL KRIL+Y+ GT+DYGI++ + LVGY D+D++G DDR
Sbjct: 4141 CARYQANPKISHLNQVKRILKYVNGTSDYGIMYCH----CSDSMLVGYCDADWAGSADDR 4308
Query: 386 KSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKK 445
KSTSG F L ISW SKKQ ++LS+ EAEYIA + Q VW++++LKE V +
Sbjct: 4309 KSTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYN-VEQD 4485
Query: 446 PMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADIFTK 505
M L DN+SAI+++KNP+ H R+KHI++R H++R+ V+ + L + T+EQ ADIFTK
Sbjct: 4486 VMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTK 4665
Query: 506 PLKTERFVILRKEIGVVSL 524
L +F LR ++G+ L
Sbjct: 4666 ALDANQFEKLRGKLGICLL 4722
>TC223792 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (7%)
Length = 804
Score = 250 bits (639), Expect = 1e-66
Identities = 126/237 (53%), Positives = 175/237 (73%)
Frame = +1
Query: 297 VDASLYKQLVGSLRFLCNSRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGI 356
VD + +++L+GSLR+LCNSR +I + V LIS+FM P+ SH+ AAKR+LR ++GT G+
Sbjct: 7 VDVTEFRRLIGSLRYLCNSRPNICFAVSLISRFMKRPRLSHMQAAKRVLRLIKGTIGSGV 186
Query: 357 LFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCE 416
LFP + K GK +L+GYTDSD+ D + KST GYLF + AP++ +SKKQ VIALS+CE
Sbjct: 187 LFPFKAKSGKP-DLLGYTDSDWKRDPEQEKSTGGYLFMYNDAPVA*SSKKQDVIALSTCE 363
Query: 417 AEYIAGSFAACQAVWLEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRF 476
AEY+A S ACQAVW+ LL+E+ +KP+ L++DN SAI+LAK+P HGRSKHIE+RF
Sbjct: 364 AEYVAASLGACQAVWMMNLLEELKLRERKPVNLLIDNKSAINLAKHPTLHGRSKHIELRF 543
Query: 477 HFLREQVNGGKLELVYCPTDEQKADIFTKPLKTERFVILRKEIGVVSLGYLN*GGVL 533
H++R+QV+ G + + YC +EQ AD+ TKP++ RF + E+ V +L LN*G VL
Sbjct: 544 HYIRDQVSKGNVTVEYCKAEEQLADLMTKPIQVSRFKQICSEL-VNNLEDLN*GRVL 711
>TC213888 similar to UP|Q9SFE1 (Q9SFE1) T26F17.17, partial (11%)
Length = 493
Score = 159 bits (401), Expect = 4e-39
Identities = 72/141 (51%), Positives = 102/141 (72%)
Frame = +3
Query: 383 DDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFV 442
DDRKST+G++F + +W SKKQP++ LS+CEAEY+A + C A+WL LLKE+
Sbjct: 9 DDRKSTTGFVFFMGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELKMP 188
Query: 443 TKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADI 502
++PME+ VDN SA++LAKNP+ H +SKHI+ R+HF+RE + +++L Y + +Q ADI
Sbjct: 189 QEEPMEICVDNKSALALAKNPVFHEKSKHIDTRYHFIRECIEKKEVKLKYVMSQDQAADI 368
Query: 503 FTKPLKTERFVILRKEIGVVS 523
FTKPLK E FV LR +GV +
Sbjct: 369 FTKPLKLETFVKLRSMLGVTN 431
>TC232995
Length = 1009
Score = 154 bits (390), Expect = 7e-38
Identities = 76/174 (43%), Positives = 113/174 (64%), Gaps = 1/174 (0%)
Frame = +2
Query: 137 VSQPPGFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKS 196
V QPPGFEI K N V+KL KALYGLKQAPRAW +R+ FLL+ +F R V+ +++++
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 197 HGDTGLLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGI 256
H D +LL+ +YVDD++ ++ E ++SEFEMS +G+L YFLG+Q + T GI
Sbjct: 182 HND--ILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GI 355
Query: 257 FMYQQKYIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEG-DVDASLYKQLVGSL 309
F+ Q KY K+++KRF M + ++TP N L +D+ G +D Y+ +G +
Sbjct: 356 FINQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEV 517
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 129 bits (324), Expect(2) = 8e-34
Identities = 72/188 (38%), Positives = 113/188 (59%), Gaps = 1/188 (0%)
Frame = +3
Query: 203 LLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQK 262
L+I +YVDD++ + + E +++K FE S G+L + LG+Q GIF++Q+K
Sbjct: 489 LIIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIFIHQEK 668
Query: 263 YIKDILKRFNMGNCHPVTTPSEVNIKLRRDDEGDVDA-SLYKQLVGSLRFLCNSRLDISY 321
Y K LKRF M P+ TP + + +D++G+ + Y ++ SL +L +SR DI +
Sbjct: 669 YTKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTSSRPDIVF 848
Query: 322 GVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGD 381
V L ++F PK SH+ A KRILRYL GTT++ + F ++ E +L+GY D ++GD
Sbjct: 849 VVCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKR----SEFDLLGYCDVYFAGD 1016
Query: 382 LDDRKSTS 389
+RKSTS
Sbjct: 1017KVERKSTS 1040
Score = 33.1 bits (74), Expect(2) = 8e-34
Identities = 16/46 (34%), Positives = 25/46 (53%)
Frame = +2
Query: 154 KLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGD 199
K +YGLKQA RAW +R+ FL+ F R + ++ + G+
Sbjct: 347 KTLSCVYGLKQALRAWYERLSSFLVSNGFTRGITDPALFRKAQKGN 484
>BU549979
Length = 615
Score = 132 bits (331), Expect = 5e-31
Identities = 68/190 (35%), Positives = 112/190 (58%), Gaps = 2/190 (1%)
Frame = -1
Query: 325 LISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDD 384
++ ++ P H AK+++RYLQGT DY +++ K LE++GY+DSD++G +D
Sbjct: 615 VLGRYQSNPGIDHWKTAKKVMRYLQGTKDYMLMY----KQTNCLEVIGYSDSDFAGCVDS 448
Query: 385 RKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFV-- 442
R+STSGY+F L +SW S KQ +IA S+ E E++ A VWL+ + + V
Sbjct: 447 RRSTSGYIFMLADGVVSWRSSKQTLIATSTMEVEFVPCFEATSHGVWLKSFMSSLRVVDS 268
Query: 443 TKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADI 502
+P++L DN +A+ +AKN S RSKHI++++ +RE+V K+ + + T+ D
Sbjct: 267 ISRPLKLYCDNFAAVFMAKNNKSGNRSKHIDIKYLVIRERVKEKKVVIEHVNTELMIVDP 88
Query: 503 FTKPLKTERF 512
TK + + F
Sbjct: 87 LTKGMTPKNF 58
>TC231899 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (30%)
Length = 687
Score = 129 bits (324), Expect = 3e-30
Identities = 65/165 (39%), Positives = 104/165 (62%)
Frame = +2
Query: 369 ELVGYTDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQ 428
+L GY D+D++G DR+STSGY + G +SW SKKQ V+A SS EAEY + + C+
Sbjct: 14 QLSGYCDADWAGCPMDRRSTSGYCVFIGGNLVSWKSKKQTVVARSSAEAEYRSMAMVTCE 193
Query: 429 AVWLEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKL 488
+W+++ L+E+ F + M+L DN +A+ +A NP+ H R+KHIE+ HF+RE++ ++
Sbjct: 194 LMWIKQFLQELRFCEELQMKLYCDNQAALHIASNPVFHERTKHIEIDCHFIREKLLSKEI 373
Query: 489 ELVYCPTDEQKADIFTKPLKTERFVILRKEIGVVSLGYLN*GGVL 533
+ +++Q DI TK L+ + I+ ++G L *GGVL
Sbjct: 374 VTEFIGSNDQPVDILTKSLRGPKIQIVCSKLGAYDLYAPA*GGVL 508
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 127 bits (319), Expect = 1e-29
Identities = 58/98 (59%), Positives = 77/98 (78%)
Frame = -3
Query: 82 FSTKPGLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPP 141
++ + G+DY E +APVAR+E IR++++ S N+ ++Q+DVKSAFLNG ++EEVYV QPP
Sbjct: 296 YNQEEGIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPP 117
Query: 142 GFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQ 179
GFEI K V+KL KALYGLKQAPRAW +RI FLL+
Sbjct: 116 GFEIPDKPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>BI321712
Length = 399
Score = 126 bits (316), Expect = 3e-29
Identities = 58/127 (45%), Positives = 92/127 (71%), Gaps = 1/127 (0%)
Frame = -3
Query: 205 ICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFMYQQKYI 264
+CLYVDDL+ TG++P +E K+ + +EFEM+D+G + Y+LG++ + GIF+ Q+ Y
Sbjct: 385 LCLYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQEDKGIFITQEGYA 206
Query: 265 KDILKRFNMGNCHPVTTPSEVNIKLRRDDEGD-VDASLYKQLVGSLRFLCNSRLDISYGV 323
K++LK+F M + +PV TP E KL + ++G+ VD +LYK L+GSLR+L +R DI Y V
Sbjct: 205 KEVLKKFKMDDANPVGTPMECGSKLSKHEKGENVDPTLYKSLIGSLRYLTCTRPDILYVV 26
Query: 324 GLISKFM 330
G++S++M
Sbjct: 25 GVVSRYM 5
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 125 bits (314), Expect = 5e-29
Identities = 71/162 (43%), Positives = 100/162 (60%), Gaps = 1/162 (0%)
Frame = +1
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G DY+ F+PVA++ + L+ S+A +WP+F LD K+AFL+G LEEEVY+ QP GF Q
Sbjct: 85 GQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFLHGYLEEEVYMEQPLGFVAQ 264
Query: 147 GK-ENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLI 205
G+ N V +L ++ YGLKQ+PRAW G + H ++ V+ H G + +
Sbjct: 265 GESSNMVCQLCRSFYGLKQSPRAWPFLYCGAAIWYDSH--EADHSVFY--CHSPQGCIYL 432
Query: 206 CLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGM 247
+YVDD+ ITGSD + LK L +F+ DLGKL YFLG+
Sbjct: 433 IVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>BG508993
Length = 374
Score = 122 bits (306), Expect = 4e-28
Identities = 56/127 (44%), Positives = 85/127 (66%)
Frame = +1
Query: 349 QGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQP 408
+GT D+G+ + +LVG+ DSD++GD+DDRKST+G++F + +W+SKKQ
Sbjct: 4 KGTIDFGLFYSPSNNY----KLVGFCDSDFAGDVDDRKSTTGFVFFMGDCVFTWSSKKQG 171
Query: 409 VIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVTKKPMELMVDNISAISLAKNPISHGR 468
++ L +CEAEY+A + C A+WL LL+E+ + K+ ++ VDN SA LAKN + H R
Sbjct: 172 IVTLFTCEAEYVAATSCTCHAIWLRRLLEELQLLQKESTKIYVDNRSAQELAKNSVFHER 351
Query: 469 SKHIEVR 475
SKHI+ R
Sbjct: 352 SKHIDTR 372
>AW201343
Length = 396
Score = 99.0 bits (245), Expect(2) = 4e-25
Identities = 50/97 (51%), Positives = 69/97 (70%)
Frame = -3
Query: 342 KRILRYLQGTTDYGILFPRQKKLGKELELVGYTDSDYSGDLDDRKSTSGYLFQLHGAPIS 401
K +L Y++G +YG+LFP K L + EL+ Y+D D G ++ +TSGY+ + APIS
Sbjct: 289 KTVLTYIKGILEYGVLFP-SKLLQQGNELIVYSDFDSCGK-SNKNNTSGYILKFLEAPIS 116
Query: 402 WTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKE 438
W SKKQPV+ALS+CEA+YIA SFA C +WL+ +LKE
Sbjct: 115 WCSKKQPVVALSTCEAKYIARSFATC*YIWLDLVLKE 5
Score = 34.3 bits (77), Expect(2) = 4e-25
Identities = 14/30 (46%), Positives = 22/30 (72%)
Frame = -1
Query: 305 LVGSLRFLCNSRLDISYGVGLISKFMCAPK 334
L+G LR+LCNSR +I++ + L+S +M K
Sbjct: 396 LIGYLRYLCNSRPNIAFSIRLVSGYMSEAK 307
>BG840035 weakly similar to GP|9294121|dbj copia-like retrotransposable
element {Arabidopsis thaliana}, partial (3%)
Length = 804
Score = 108 bits (271), Expect = 5e-24
Identities = 56/139 (40%), Positives = 85/139 (60%), Gaps = 1/139 (0%)
Frame = +2
Query: 384 DRKSTSGYLFQ-LHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFV 442
D KS +G LF L W+SKKQ EAEY+A + A QA+WL +L +
Sbjct: 146 DIKSXTGLLFSALVRGRFFWSSKKQDX------EAEYVATTTAVNQAIWLRRILANLHME 307
Query: 443 TKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCPTDEQKADI 502
K+P +++VDN + IS++ N + HGR+KH +++ FLRE G+++L+YC +++Q ADI
Sbjct: 308 QKEPTQILVDNQTTISISNNHVFHGRTKHFKIKLFFLRETQREGEVKLIYCRSEDQSADI 487
Query: 503 FTKPLKTERFVILRKEIGV 521
TK RF +LR ++GV
Sbjct: 488 LTKVFPKVRFEVLRNKLGV 544
>AW185460
Length = 411
Score = 108 bits (271), Expect = 5e-24
Identities = 55/107 (51%), Positives = 69/107 (64%)
Frame = +2
Query: 315 SRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKLGKELELVGYT 374
+R DI Y L+S+FM +P + H A KRILRYLQGT +GI + + EL+GYT
Sbjct: 92 TRPDIMYATSLLSRFMQSPSQIHFGAGKRILRYLQGTKAFGIWYTTETNS----ELLGYT 259
Query: 375 DSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIA 421
DSD++G DD KSTSGY F L SW SKKQ +A S+ EAEY+A
Sbjct: 260 DSDWAGSTDDMKSTSGYAFSLGSGMFSWASKKQATVAQSTAEAEYVA 400
>BM143109
Length = 415
Score = 107 bits (267), Expect = 1e-23
Identities = 57/137 (41%), Positives = 84/137 (60%)
Frame = +1
Query: 139 QPPGFEIQGKENSVFKLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHG 198
QPP + K N VFKL K LYGLKQA RAW + + FLL F + V+ +++ K
Sbjct: 4 QPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLN 183
Query: 199 DTGLLLICLYVDDLLITGSDPKELDELKQILKSEFEMSDLGKLGYFLGMQFEYTAAGIFM 258
D +LL+ +YVDD++ ++ + Q +++EFEMS + +L +FLG+Q + T GIF+
Sbjct: 184 D--ILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFI 357
Query: 259 YQQKYIKDILKRFNMGN 275
Q KY KD++ RF M N
Sbjct: 358 SQSKYCKDLIHRFGMEN 408
>CO983516
Length = 724
Score = 107 bits (266), Expect = 2e-23
Identities = 54/120 (45%), Positives = 81/120 (67%)
Frame = +2
Query: 94 FAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQGKENSVF 153
F PVAR+E+IRL++ +A + ++Q+DVKSAFLNG L EEVYV QP GF + V+
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 154 KLSKALYGLKQAPRAWNKRIDGFLLQLKFHRCSVEYGVYVRKSHGDTGLLLICLYVDDLL 213
+L KALYGLKQAPRAW +R+ L Q + + ++ ++V++ L++ +YVDD++
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQD--AENLMIAQIYVDDIV 718
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 107 bits (266), Expect = 2e-23
Identities = 45/83 (54%), Positives = 67/83 (80%)
Frame = -2
Query: 87 GLDYSEVFAPVARIETIRLVVSIASARNWPIFQLDVKSAFLNGPLEEEVYVSQPPGFEIQ 146
G+DY + F+PVA++ T+RL +++A+ +WP+ QLD+K+AFL+G LEE++Y+ QPPGF Q
Sbjct: 280 GIDYCDTFSPVAKLTTVRLFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQ 101
Query: 147 GKENSVFKLSKALYGLKQAPRAW 169
G+ V KL ++LYGLKQ+PRAW
Sbjct: 100 GEYGLVCKLHRSLYGLKQSPRAW 32
>TC213445
Length = 705
Score = 76.6 bits (187), Expect(2) = 3e-23
Identities = 42/97 (43%), Positives = 59/97 (60%)
Frame = +1
Query: 384 DRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEELLKEMGFVT 443
DR+STS + A +SW SKKQ + LS+ EAEYI+ Q W+ + L + G +
Sbjct: 406 DRESTSDTCHFIGSALVSWHSKKQNSVVLSTAEAEYISARSYYAQIFWMRQQLFDYG-LK 582
Query: 444 KKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLR 480
+ + DN SAI+L+KN I + R+KHIE+R HFLR
Sbjct: 583 LDHIPIRCDNTSAINLSKNHILYSRTKHIEIRHHFLR 693
Score = 50.4 bits (119), Expect(2) = 3e-23
Identities = 25/72 (34%), Positives = 41/72 (56%)
Frame = +2
Query: 305 LVGSLRFLCNSRLDISYGVGLISKFMCAPKKSHLAAAKRILRYLQGTTDYGILFPRQKKL 364
++ S +L SR I + V + ++ PK+SHL+ KRI+RYL G + G+ +P+
Sbjct: 197 MIESFLYLSTSRPHIMFSVCMCVRYQANPKESHLSVIKRIMRYLLGIINLGLWYPK---- 364
Query: 365 GKELELVGYTDS 376
LVGY+D+
Sbjct: 365 NSSYNLVGYSDA 400
>BU548243
Length = 599
Score = 100 bits (250), Expect = 1e-21
Identities = 54/147 (36%), Positives = 88/147 (59%)
Frame = -1
Query: 375 DSDYSGDLDDRKSTSGYLFQLHGAPISWTSKKQPVIALSSCEAEYIAGSFAACQAVWLEE 434
D+ ++ D+DD +ST G L ISW S+KQ V A SS EAEY + + + + W++
Sbjct: 587 DAGWASDVDDHRSTLGSAIFLGPNLISWWSRKQQVTAQSSTEAEYRSIAQTSAELTWIQA 408
Query: 435 LLKEMGFVTKKPMELMVDNISAISLAKNPISHGRSKHIEVRFHFLREQVNGGKLELVYCP 494
LL E+ P+ ++ DN SA+++A N + H R+KH+E+ F+ E+V +L++ + P
Sbjct: 407 LLMELQIPFTPPV-ILCDNKSAVAIAHNLVFHSRTKHMEIDVFFVHEKVLSKQLQIFHIP 231
Query: 495 TDEQKADIFTKPLKTERFVILRKEIGV 521
+Q A I TKPL + RF L+ ++ V
Sbjct: 230 ALDQWAGILTKPLSSARFTFLKSKLTV 150
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.331 0.146 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,634,480
Number of Sequences: 63676
Number of extensions: 299945
Number of successful extensions: 1503
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 1460
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1462
length of query: 533
length of database: 12,639,632
effective HSP length: 102
effective length of query: 431
effective length of database: 6,144,680
effective search space: 2648357080
effective search space used: 2648357080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0028b.2


